Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6554
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7545 | 3.476e-03 | 5.640e-04 | 7.691e-01 | 1.508e-06 |
|---|
| Loi | 0.2306 | 7.980e-02 | 1.124e-02 | 4.271e-01 | 3.830e-04 |
|---|
| Schmidt | 0.6729 | 0.000e+00 | 0.000e+00 | 4.550e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.462e-03 | 0.000e+00 |
|---|
| Wang | 0.2617 | 2.667e-03 | 4.372e-02 | 3.959e-01 | 4.616e-05 |
|---|
Expression data for subnetwork 6554 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
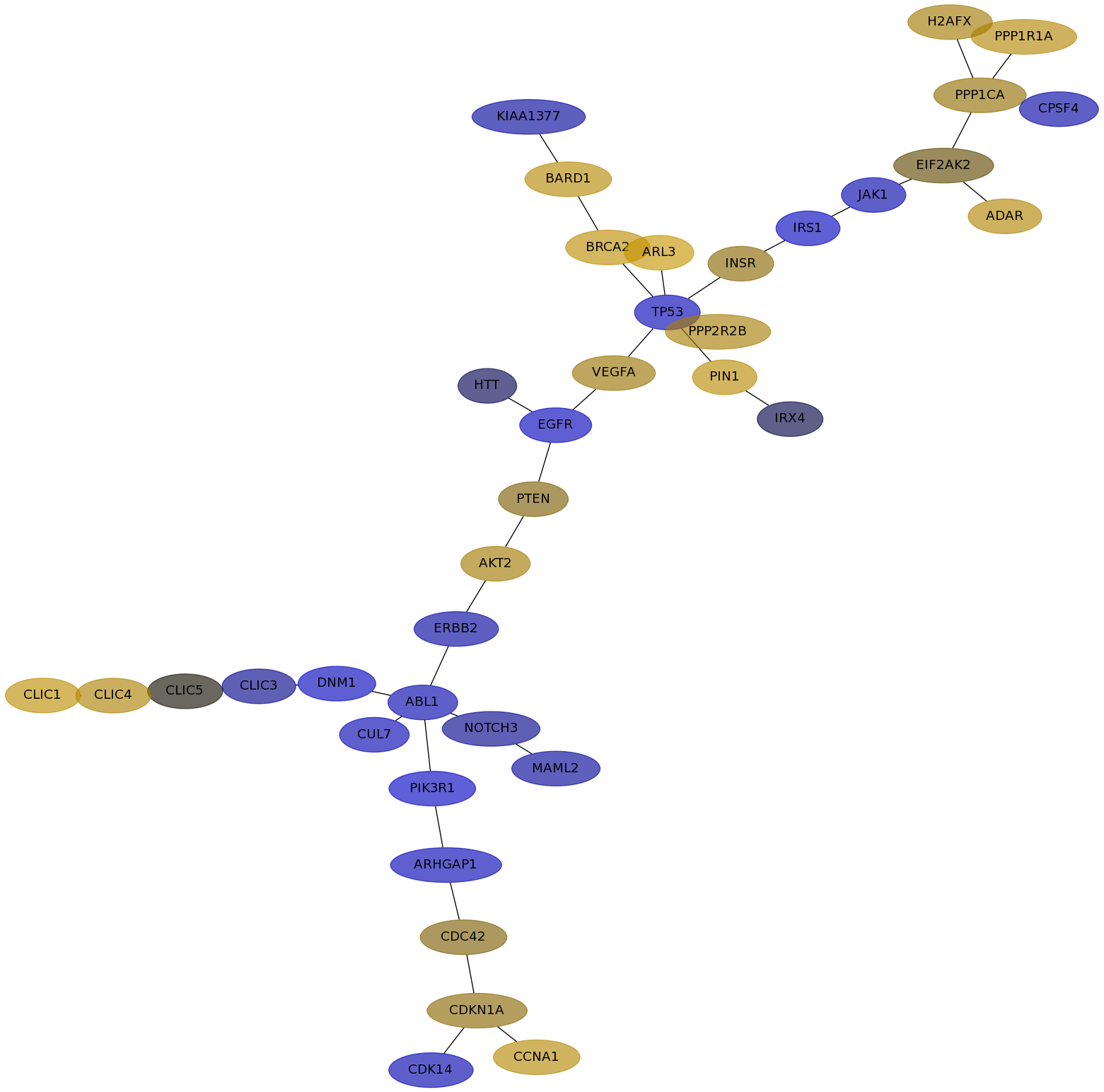
Score for each gene in subnetwork 6554 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| CLIC5 |   | 1 | 1195 | 1621 | 1637 | 0.002 | 0.027 | 0.109 | -0.284 | -0.133 |
|---|
| CUL7 |   | 3 | 557 | 1621 | 1593 | -0.157 | 0.176 | -0.108 | -0.006 | 0.104 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| IRX4 |   | 2 | 743 | 412 | 439 | -0.012 | 0.137 | -0.018 | -0.139 | 0.125 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| KIAA1377 |   | 18 | 86 | 141 | 136 | -0.095 | 0.116 | undef | 0.198 | undef |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| CDK14 |   | 7 | 256 | 179 | 201 | -0.151 | 0.011 | -0.071 | 0.212 | 0.107 |
|---|
| ARL3 |   | 6 | 301 | 83 | 105 | 0.266 | -0.002 | 0.203 | 0.206 | -0.029 |
|---|
| CLIC3 |   | 1 | 1195 | 1621 | 1637 | -0.061 | 0.158 | -0.255 | -0.053 | -0.053 |
|---|
| ABL1 |   | 13 | 124 | 727 | 699 | -0.152 | 0.200 | 0.108 | undef | -0.059 |
|---|
| CLIC4 |   | 1 | 1195 | 1621 | 1637 | 0.141 | 0.196 | 0.282 | -0.040 | 0.106 |
|---|
| EIF2AK2 |   | 9 | 196 | 648 | 629 | 0.023 | -0.022 | -0.134 | 0.096 | -0.303 |
|---|
| CDC42 |   | 26 | 52 | 179 | 177 | 0.047 | 0.242 | -0.027 | 0.040 | -0.042 |
|---|
| PTEN |   | 27 | 50 | 141 | 135 | 0.045 | 0.055 | 0.017 | 0.183 | -0.041 |
|---|
| PIN1 |   | 24 | 62 | 179 | 181 | 0.194 | 0.040 | -0.040 | 0.065 | -0.048 |
|---|
| ARHGAP1 |   | 6 | 301 | 179 | 205 | -0.167 | 0.297 | 0.001 | -0.032 | 0.251 |
|---|
| CLIC1 |   | 1 | 1195 | 1621 | 1637 | 0.208 | 0.060 | -0.019 | 0.099 | 0.178 |
|---|
| ADAR |   | 2 | 743 | 1294 | 1286 | 0.154 | -0.104 | -0.127 | 0.174 | -0.169 |
|---|
| NOTCH3 |   | 5 | 360 | 806 | 784 | -0.063 | 0.115 | 0.136 | undef | -0.082 |
|---|
| PPP1CA |   | 2 | 743 | 1621 | 1611 | 0.076 | -0.061 | 0.020 | 0.196 | -0.168 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| CPSF4 |   | 1 | 1195 | 1621 | 1637 | -0.114 | 0.007 | -0.013 | 0.060 | -0.098 |
|---|
| DNM1 |   | 4 | 440 | 236 | 262 | -0.203 | 0.095 | 0.163 | 0.001 | 0.082 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| MAML2 |   | 1 | 1195 | 1621 | 1637 | -0.088 | 0.157 | undef | undef | undef |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| PPP1R1A |   | 2 | 743 | 1 | 66 | 0.165 | -0.061 | 0.171 | 0.353 | 0.115 |
|---|
| IRS1 |   | 9 | 196 | 842 | 805 | -0.219 | 0.065 | 0.165 | 0.102 | 0.064 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| JAK1 |   | 5 | 360 | 696 | 680 | -0.140 | 0.034 | -0.176 | 0.111 | 0.159 |
|---|
| HTT |   | 14 | 117 | 141 | 140 | -0.016 | -0.015 | 0.075 | 0.092 | -0.093 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| H2AFX |   | 7 | 256 | 179 | 201 | 0.111 | -0.072 | 0.121 | 0.165 | -0.010 |
|---|
GO Enrichment output for subnetwork 6554 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 3.586E-10 | 8.761E-07 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 1.073E-09 | 1.311E-06 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 1.229E-09 | 1.001E-06 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 2.499E-09 | 1.526E-06 |
|---|
| negative regulation of cell migration | GO:0030336 |  | 4.894E-09 | 2.391E-06 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 4.987E-09 | 2.03E-06 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 4.987E-09 | 1.74E-06 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 4.987E-09 | 1.523E-06 |
|---|
| negative regulation of cell motion | GO:0051271 |  | 1.482E-08 | 4.024E-06 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 1.489E-08 | 3.638E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.847E-08 | 4.102E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 4.633E-10 | 1.115E-06 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 1.387E-09 | 1.668E-06 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 1.686E-09 | 1.352E-06 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 3.228E-09 | 1.941E-06 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 3.228E-09 | 1.553E-06 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 6.44E-09 | 2.582E-06 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 6.44E-09 | 2.213E-06 |
|---|
| negative regulation of cell migration | GO:0030336 |  | 7.101E-09 | 2.136E-06 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 1.923E-08 | 5.14E-06 |
|---|
| chloride transport | GO:0006821 |  | 2.148E-08 | 5.168E-06 |
|---|
| negative regulation of cell motion | GO:0051271 |  | 2.148E-08 | 4.698E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 6.429E-09 | 1.479E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.282E-08 | 1.474E-05 |
|---|
| chloride transport | GO:0006821 |  | 2.487E-08 | 1.907E-05 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 3.825E-08 | 2.199E-05 |
|---|
| inorganic anion transport | GO:0015698 |  | 2.091E-07 | 9.62E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.905E-07 | 1.497E-04 |
|---|
| nuclear migration | GO:0007097 |  | 5.112E-07 | 1.68E-04 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 5.112E-07 | 1.47E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 5.112E-07 | 1.306E-04 |
|---|
| negative regulation of cell migration | GO:0030336 |  | 6.371E-07 | 1.465E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 7.146E-07 | 1.494E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cell migration | GO:0030334 |  | 1.991E-08 | 3.669E-05 |
|---|
| regulation of locomotion | GO:0040012 |  | 4.234E-08 | 3.902E-05 |
|---|
| regulation of cell motion | GO:0051270 |  | 4.234E-08 | 2.601E-05 |
|---|
| chloride transport | GO:0006821 |  | 9.976E-08 | 4.596E-05 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.211E-07 | 4.462E-05 |
|---|
| inorganic anion transport | GO:0015698 |  | 3.911E-07 | 1.201E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 4.528E-07 | 1.192E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 6.85E-07 | 1.578E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.642E-06 | 3.362E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.642E-06 | 3.026E-04 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 2.1E-06 | 3.519E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 6.429E-09 | 1.479E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.282E-08 | 1.474E-05 |
|---|
| chloride transport | GO:0006821 |  | 2.487E-08 | 1.907E-05 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 3.825E-08 | 2.199E-05 |
|---|
| inorganic anion transport | GO:0015698 |  | 2.091E-07 | 9.62E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.905E-07 | 1.497E-04 |
|---|
| nuclear migration | GO:0007097 |  | 5.112E-07 | 1.68E-04 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 5.112E-07 | 1.47E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 5.112E-07 | 1.306E-04 |
|---|
| negative regulation of cell migration | GO:0030336 |  | 6.371E-07 | 1.465E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 7.146E-07 | 1.494E-04 |
|---|



