Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6536
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7549 | 3.451e-03 | 5.590e-04 | 7.680e-01 | 1.482e-06 |
|---|
| Loi | 0.2305 | 8.002e-02 | 1.129e-02 | 4.276e-01 | 3.864e-04 |
|---|
| Schmidt | 0.6727 | 0.000e+00 | 0.000e+00 | 4.563e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.472e-03 | 0.000e+00 |
|---|
| Wang | 0.2615 | 2.694e-03 | 4.396e-02 | 3.970e-01 | 4.702e-05 |
|---|
Expression data for subnetwork 6536 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
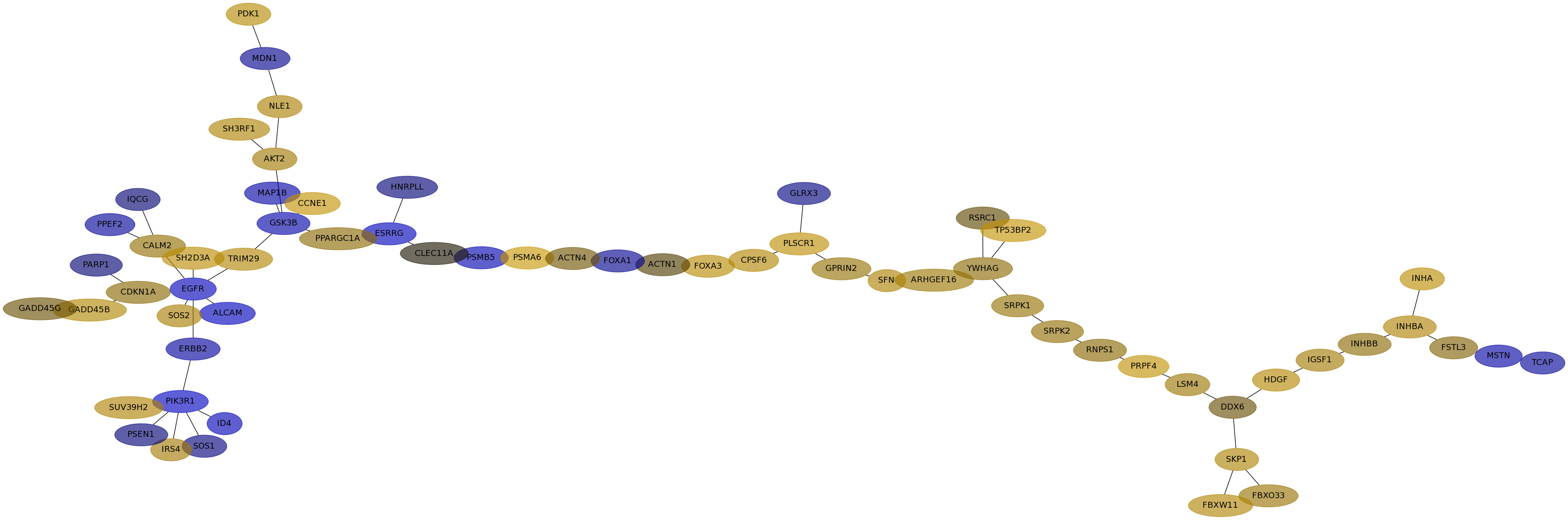
Score for each gene in subnetwork 6536 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| FBXW11 |   | 3 | 557 | 658 | 654 | 0.154 | 0.225 | 0.262 | undef | 0.169 |
|---|
| PSMB5 |   | 2 | 743 | 692 | 713 | -0.163 | 0.195 | undef | 0.137 | undef |
|---|
| SH2D3A |   | 3 | 557 | 1510 | 1486 | 0.197 | 0.141 | -0.106 | -0.012 | 0.121 |
|---|
| HDGF |   | 1 | 1195 | 1752 | 1757 | 0.171 | -0.106 | -0.048 | 0.015 | -0.055 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| FOXA3 |   | 12 | 140 | 318 | 322 | 0.192 | 0.289 | undef | -0.171 | undef |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| LSM4 |   | 1 | 1195 | 1752 | 1757 | 0.098 | -0.124 | -0.038 | 0.063 | -0.057 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PSEN1 |   | 10 | 167 | 366 | 362 | -0.041 | 0.110 | undef | 0.083 | undef |
|---|
| GLRX3 |   | 1 | 1195 | 1752 | 1757 | -0.053 | 0.027 | -0.088 | -0.057 | -0.096 |
|---|
| NLE1 |   | 3 | 557 | 882 | 871 | 0.138 | -0.086 | -0.018 | 0.001 | 0.044 |
|---|
| RSRC1 |   | 21 | 73 | 366 | 354 | 0.023 | 0.104 | 0.105 | -0.098 | 0.212 |
|---|
| HNRPLL |   | 1 | 1195 | 1752 | 1757 | -0.041 | -0.070 | undef | undef | undef |
|---|
| CCNE1 |   | 4 | 440 | 1083 | 1067 | 0.245 | -0.141 | -0.077 | undef | -0.018 |
|---|
| IGSF1 |   | 1 | 1195 | 1752 | 1757 | 0.117 | 0.024 | 0.056 | 0.068 | -0.107 |
|---|
| ARHGEF16 |   | 10 | 167 | 296 | 283 | 0.096 | 0.036 | -0.047 | 0.200 | 0.038 |
|---|
| CALM2 |   | 2 | 743 | 957 | 958 | 0.076 | 0.056 | 0.104 | -0.004 | 0.086 |
|---|
| IQCG |   | 1 | 1195 | 1752 | 1757 | -0.036 | 0.041 | undef | undef | undef |
|---|
| PSMA6 |   | 5 | 360 | 842 | 828 | 0.280 | -0.118 | -0.018 | undef | -0.091 |
|---|
| SUV39H2 |   | 8 | 222 | 318 | 325 | 0.150 | -0.097 | -0.057 | undef | 0.028 |
|---|
| INHBB |   | 1 | 1195 | 1752 | 1757 | 0.066 | 0.299 | 0.065 | 0.185 | 0.165 |
|---|
| DDX6 |   | 4 | 440 | 1688 | 1656 | 0.028 | 0.162 | 0.157 | -0.035 | 0.084 |
|---|
| GPRIN2 |   | 16 | 104 | 179 | 190 | 0.084 | 0.078 | 0.012 | 0.054 | 0.114 |
|---|
| FBXO33 |   | 1 | 1195 | 1752 | 1757 | 0.090 | -0.072 | undef | undef | undef |
|---|
| TCAP |   | 1 | 1195 | 1752 | 1757 | -0.089 | 0.061 | -0.020 | 0.064 | -0.029 |
|---|
| TP53BP2 |   | 11 | 148 | 692 | 668 | 0.245 | -0.012 | 0.129 | 0.021 | 0.017 |
|---|
| CPSF6 |   | 6 | 301 | 318 | 330 | 0.164 | 0.052 | 0.091 | 0.129 | -0.088 |
|---|
| INHBA |   | 2 | 743 | 1752 | 1744 | 0.151 | 0.188 | 0.037 | 0.150 | 0.143 |
|---|
| SRPK2 |   | 3 | 557 | 1203 | 1187 | 0.076 | 0.183 | -0.087 | undef | 0.054 |
|---|
| SOS2 |   | 1 | 1195 | 1752 | 1757 | 0.128 | 0.098 | -0.112 | undef | 0.036 |
|---|
| CLEC11A |   | 7 | 256 | 692 | 673 | 0.004 | 0.353 | 0.005 | 0.081 | 0.272 |
|---|
| PLSCR1 |   | 10 | 167 | 412 | 408 | 0.202 | -0.159 | 0.091 | undef | -0.105 |
|---|
| PDK1 |   | 10 | 167 | 1 | 40 | 0.183 | -0.138 | -0.041 | -0.265 | -0.126 |
|---|
| PARP1 |   | 29 | 45 | 83 | 84 | -0.036 | -0.162 | -0.089 | 0.238 | 0.012 |
|---|
| MSTN |   | 1 | 1195 | 1752 | 1757 | -0.113 | 0.055 | -0.118 | 0.103 | 0.153 |
|---|
| SOS1 |   | 5 | 360 | 478 | 476 | -0.049 | 0.002 | -0.007 | 0.160 | -0.042 |
|---|
| ESRRG |   | 6 | 301 | 614 | 602 | -0.205 | 0.197 | -0.007 | 0.155 | -0.085 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| RNPS1 |   | 1 | 1195 | 1752 | 1757 | 0.063 | -0.151 | -0.083 | 0.031 | 0.042 |
|---|
| ACTN1 |   | 18 | 86 | 318 | 313 | 0.016 | 0.347 | 0.068 | 0.100 | 0.314 |
|---|
| INHA |   | 1 | 1195 | 1752 | 1757 | 0.200 | -0.058 | -0.091 | -0.055 | 0.153 |
|---|
| PPEF2 |   | 1 | 1195 | 1752 | 1757 | -0.089 | -0.184 | -0.088 | 0.220 | -0.092 |
|---|
| ALCAM |   | 8 | 222 | 1 | 46 | -0.208 | 0.193 | 0.067 | 0.088 | -0.010 |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| YWHAG |   | 24 | 62 | 296 | 279 | 0.067 | 0.024 | undef | 0.266 | undef |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| SH3RF1 |   | 6 | 301 | 1222 | 1194 | 0.144 | 0.238 | undef | undef | undef |
|---|
| GADD45B |   | 8 | 222 | 318 | 325 | 0.158 | 0.122 | 0.067 | 0.060 | 0.212 |
|---|
| FSTL3 |   | 1 | 1195 | 1752 | 1757 | 0.051 | 0.155 | 0.001 | 0.078 | 0.003 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| ACTN4 |   | 14 | 117 | 318 | 315 | 0.035 | 0.180 | undef | -0.048 | undef |
|---|
| SKP1 |   | 2 | 743 | 727 | 734 | 0.143 | 0.037 | 0.055 | 0.148 | -0.110 |
|---|
| PRPF4 |   | 1 | 1195 | 1752 | 1757 | 0.232 | -0.079 | 0.196 | 0.133 | -0.045 |
|---|
| MDN1 |   | 1 | 1195 | 1752 | 1757 | -0.070 | -0.071 | 0.124 | undef | 0.239 |
|---|
| PPARGC1A |   | 1 | 1195 | 1752 | 1757 | 0.066 | -0.123 | 0.243 | 0.157 | 0.112 |
|---|
| SFN |   | 25 | 59 | 478 | 454 | 0.141 | 0.270 | 0.023 | -0.102 | 0.101 |
|---|
| FOXA1 |   | 27 | 50 | 179 | 176 | -0.072 | 0.071 | -0.015 | 0.214 | 0.005 |
|---|
| SRPK1 |   | 2 | 743 | 1203 | 1206 | 0.085 | -0.010 | 0.143 | 0.037 | 0.191 |
|---|
| TRIM29 |   | 8 | 222 | 296 | 287 | 0.161 | 0.240 | -0.150 | -0.077 | 0.024 |
|---|
GO Enrichment output for subnetwork 6536 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.263E-08 | 7.972E-05 |
|---|
| negative regulation of cytokine biosynthetic process | GO:0042036 |  | 3.406E-07 | 4.161E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.869E-07 | 3.151E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 4.218E-07 | 2.576E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 5.88E-07 | 2.873E-04 |
|---|
| negative regulation of hormone secretion | GO:0046888 |  | 5.902E-07 | 2.403E-04 |
|---|
| hemoglobin biosynthetic process | GO:0042541 |  | 6.473E-07 | 2.259E-04 |
|---|
| cellular glucose homeostasis | GO:0001678 |  | 6.473E-07 | 1.977E-04 |
|---|
| negative regulation of macrophage differentiation | GO:0045650 |  | 6.473E-07 | 1.757E-04 |
|---|
| positive regulation of hormone secretion | GO:0046887 |  | 9.551E-07 | 2.333E-04 |
|---|
| activation of MAPKKK activity | GO:0000185 |  | 1.291E-06 | 2.867E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.268E-08 | 1.027E-04 |
|---|
| negative regulation of cytokine biosynthetic process | GO:0042036 |  | 4.097E-07 | 4.929E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 4.227E-07 | 3.39E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 4.62E-07 | 2.779E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 6.504E-07 | 3.13E-04 |
|---|
| negative regulation of hormone secretion | GO:0046888 |  | 7.097E-07 | 2.846E-04 |
|---|
| hemoglobin biosynthetic process | GO:0042541 |  | 7.452E-07 | 2.561E-04 |
|---|
| cellular glucose homeostasis | GO:0001678 |  | 7.452E-07 | 2.241E-04 |
|---|
| negative regulation of macrophage differentiation | GO:0045650 |  | 7.452E-07 | 1.992E-04 |
|---|
| positive regulation of hormone secretion | GO:0046887 |  | 1.148E-06 | 2.763E-04 |
|---|
| activation of MAPKKK activity | GO:0000185 |  | 1.486E-06 | 3.25E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.257E-08 | 1.439E-04 |
|---|
| negative regulation of cytokine biosynthetic process | GO:0042036 |  | 3.773E-07 | 4.339E-04 |
|---|
| negative regulation of hormone secretion | GO:0046888 |  | 7.149E-07 | 5.481E-04 |
|---|
| positive regulation of hormone secretion | GO:0046887 |  | 1.237E-06 | 7.114E-04 |
|---|
| activation of MAPKKK activity | GO:0000185 |  | 2.274E-06 | 1.046E-03 |
|---|
| negative regulation of B cell activation | GO:0050869 |  | 2.274E-06 | 8.719E-04 |
|---|
| ovarian follicle development | GO:0001541 |  | 3.065E-06 | 1.007E-03 |
|---|
| hemoglobin metabolic process | GO:0020027 |  | 3.966E-06 | 1.14E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.076E-06 | 1.042E-03 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 4.5E-06 | 1.035E-03 |
|---|
| interphase | GO:0051325 |  | 4.662E-06 | 9.747E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of cytokine biosynthetic process | GO:0042036 |  | 1.089E-07 | 2.006E-04 |
|---|
| positive regulation of hormone secretion | GO:0046887 |  | 2.828E-07 | 2.606E-04 |
|---|
| negative regulation of lymphocyte activation | GO:0051250 |  | 2.828E-07 | 1.737E-04 |
|---|
| negative regulation of hormone secretion | GO:0046888 |  | 6.078E-07 | 2.8E-04 |
|---|
| negative regulation of leukocyte activation | GO:0002695 |  | 6.078E-07 | 2.24E-04 |
|---|
| ovarian follicle development | GO:0001541 |  | 1.151E-06 | 3.535E-04 |
|---|
| negative regulation of cell activation | GO:0050866 |  | 1.151E-06 | 3.03E-04 |
|---|
| hemoglobin biosynthetic process | GO:0042541 |  | 1.671E-06 | 3.85E-04 |
|---|
| cellular glucose homeostasis | GO:0001678 |  | 1.671E-06 | 3.422E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.714E-06 | 3.16E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.966E-06 | 3.294E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.257E-08 | 1.439E-04 |
|---|
| negative regulation of cytokine biosynthetic process | GO:0042036 |  | 3.773E-07 | 4.339E-04 |
|---|
| negative regulation of hormone secretion | GO:0046888 |  | 7.149E-07 | 5.481E-04 |
|---|
| positive regulation of hormone secretion | GO:0046887 |  | 1.237E-06 | 7.114E-04 |
|---|
| activation of MAPKKK activity | GO:0000185 |  | 2.274E-06 | 1.046E-03 |
|---|
| negative regulation of B cell activation | GO:0050869 |  | 2.274E-06 | 8.719E-04 |
|---|
| ovarian follicle development | GO:0001541 |  | 3.065E-06 | 1.007E-03 |
|---|
| hemoglobin metabolic process | GO:0020027 |  | 3.966E-06 | 1.14E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.076E-06 | 1.042E-03 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 4.5E-06 | 1.035E-03 |
|---|
| interphase | GO:0051325 |  | 4.662E-06 | 9.747E-04 |
|---|



