Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6534
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7549 | 3.450e-03 | 5.580e-04 | 7.680e-01 | 1.478e-06 |
|---|
| Loi | 0.2304 | 8.015e-02 | 1.133e-02 | 4.279e-01 | 3.886e-04 |
|---|
| Schmidt | 0.6728 | 0.000e+00 | 0.000e+00 | 4.557e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.473e-03 | 0.000e+00 |
|---|
| Wang | 0.2614 | 2.696e-03 | 4.398e-02 | 3.971e-01 | 4.709e-05 |
|---|
Expression data for subnetwork 6534 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
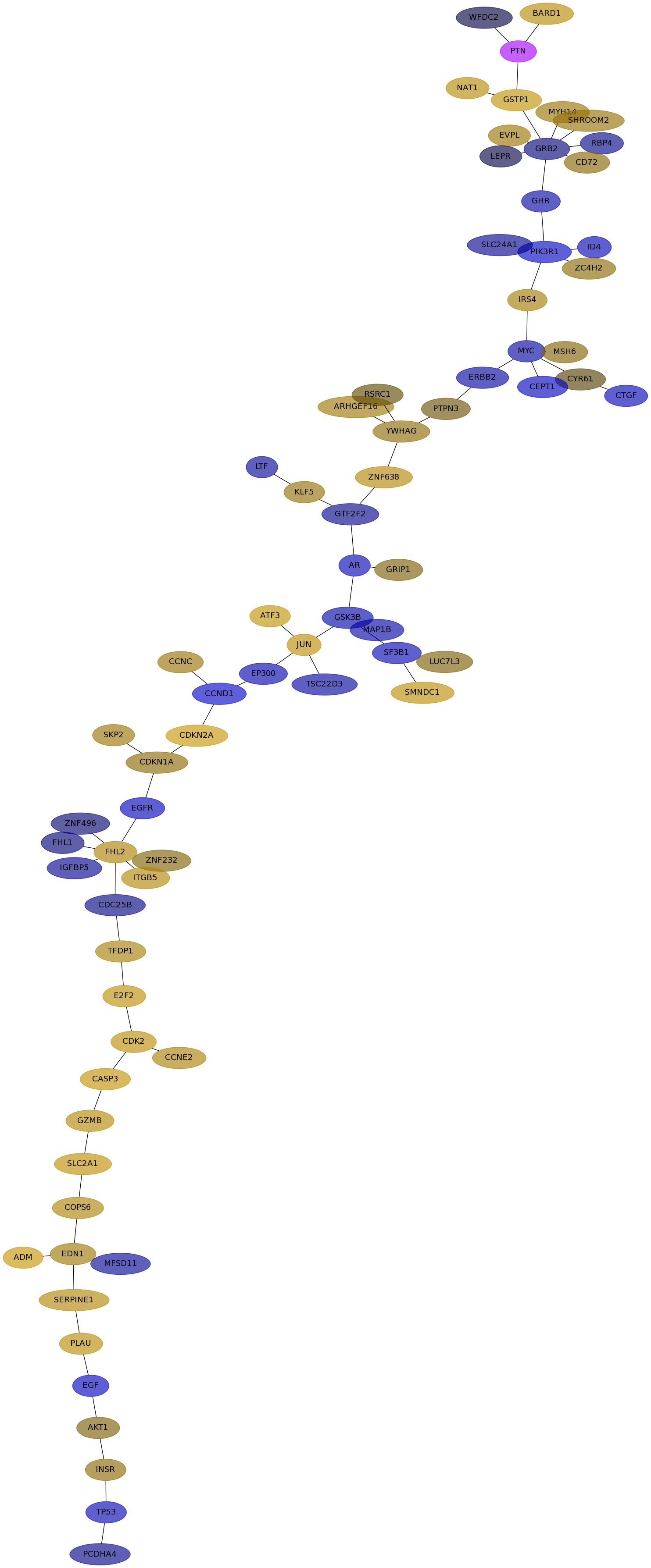
Score for each gene in subnetwork 6534 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| CCNC |   | 5 | 360 | 570 | 562 | 0.085 | -0.053 | undef | 0.028 | undef |
|---|
| CCNE2 |   | 11 | 148 | 412 | 407 | 0.136 | -0.178 | 0.241 | undef | -0.055 |
|---|
| CASP3 |   | 11 | 148 | 141 | 143 | 0.234 | -0.032 | -0.142 | -0.097 | -0.084 |
|---|
| ZNF496 |   | 1 | 1195 | 1786 | 1800 | -0.032 | -0.134 | undef | -0.044 | undef |
|---|
| SF3B1 |   | 18 | 86 | 296 | 280 | -0.154 | 0.126 | -0.073 | -0.131 | -0.204 |
|---|
| TSC22D3 |   | 1 | 1195 | 1786 | 1800 | -0.105 | 0.031 | -0.076 | undef | -0.021 |
|---|
| SLC2A1 |   | 7 | 256 | 236 | 237 | 0.205 | 0.143 | -0.050 | -0.023 | 0.086 |
|---|
| EGF |   | 12 | 140 | 638 | 620 | -0.212 | 0.182 | -0.276 | -0.060 | 0.005 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| MFSD11 |   | 1 | 1195 | 1786 | 1800 | -0.083 | -0.073 | 0.076 | undef | 0.064 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| GZMB |   | 1 | 1195 | 1786 | 1800 | 0.171 | -0.238 | -0.152 | -0.161 | -0.192 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| LTF |   | 3 | 557 | 1222 | 1210 | -0.082 | 0.060 | -0.268 | -0.081 | -0.280 |
|---|
| ARHGEF16 |   | 10 | 167 | 296 | 283 | 0.096 | 0.036 | -0.047 | 0.200 | 0.038 |
|---|
| CCND1 |   | 26 | 52 | 1 | 24 | -0.258 | 0.074 | 0.117 | undef | 0.003 |
|---|
| WFDC2 |   | 5 | 360 | 236 | 242 | -0.011 | -0.011 | 0.333 | -0.162 | -0.157 |
|---|
| PLAU |   | 12 | 140 | 179 | 192 | 0.187 | 0.153 | 0.141 | 0.072 | 0.192 |
|---|
| CTGF |   | 32 | 40 | 318 | 290 | -0.169 | 0.181 | 0.126 | 0.101 | 0.209 |
|---|
| CDC25B |   | 2 | 743 | 1771 | 1754 | -0.054 | -0.075 | -0.037 | undef | 0.009 |
|---|
| MYH14 |   | 2 | 743 | 1786 | 1776 | 0.092 | 0.103 | -0.105 | undef | -0.118 |
|---|
| ZNF638 |   | 5 | 360 | 806 | 784 | 0.164 | 0.088 | 0.020 | 0.077 | -0.045 |
|---|
| AKT1 |   | 16 | 104 | 236 | 227 | 0.045 | 0.126 | -0.099 | undef | -0.168 |
|---|
| ATF3 |   | 17 | 95 | 179 | 189 | 0.225 | -0.021 | 0.000 | undef | -0.039 |
|---|
| ADM |   | 10 | 167 | 682 | 653 | 0.255 | 0.187 | 0.091 | 0.169 | 0.212 |
|---|
| YWHAG |   | 24 | 62 | 296 | 279 | 0.067 | 0.024 | undef | 0.266 | undef |
|---|
| RBP4 |   | 4 | 440 | 764 | 761 | -0.059 | -0.011 | 0.206 | 0.334 | -0.045 |
|---|
| ZNF232 |   | 3 | 557 | 882 | 871 | 0.048 | -0.040 | 0.189 | -0.007 | 0.159 |
|---|
| SLC24A1 |   | 4 | 440 | 1323 | 1295 | -0.068 | 0.109 | 0.178 | -0.020 | 0.056 |
|---|
| LUC7L3 |   | 16 | 104 | 296 | 281 | 0.043 | 0.237 | 0.175 | 0.135 | 0.074 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| SERPINE1 |   | 8 | 222 | 179 | 200 | 0.160 | 0.200 | 0.056 | 0.070 | 0.030 |
|---|
| COPS6 |   | 16 | 104 | 1 | 31 | 0.147 | -0.068 | 0.128 | 0.216 | 0.065 |
|---|
| E2F2 |   | 13 | 124 | 1 | 35 | 0.212 | -0.047 | 0.162 | -0.131 | -0.092 |
|---|
| SMNDC1 |   | 3 | 557 | 1083 | 1073 | 0.199 | 0.079 | 0.288 | 0.002 | 0.174 |
|---|
| PTN |   | 24 | 62 | 236 | 225 | undef | 0.022 | 0.000 | 0.054 | -0.092 |
|---|
| PCDHA4 |   | 2 | 743 | 1510 | 1502 | -0.060 | 0.110 | -0.192 | 0.290 | -0.029 |
|---|
| EVPL |   | 1 | 1195 | 1786 | 1800 | 0.088 | -0.005 | -0.045 | -0.031 | -0.013 |
|---|
| PTPN3 |   | 11 | 148 | 658 | 635 | 0.029 | 0.060 | -0.067 | 0.225 | -0.034 |
|---|
| ITGB5 |   | 4 | 440 | 412 | 419 | 0.163 | 0.208 | 0.081 | 0.278 | 0.148 |
|---|
| GSTP1 |   | 14 | 117 | 1 | 34 | 0.222 | -0.166 | 0.201 | 0.126 | -0.136 |
|---|
| RSRC1 |   | 21 | 73 | 366 | 354 | 0.023 | 0.104 | 0.105 | -0.098 | 0.212 |
|---|
| JUN |   | 26 | 52 | 179 | 177 | 0.200 | 0.051 | 0.220 | undef | 0.200 |
|---|
| CYR61 |   | 4 | 440 | 991 | 978 | 0.018 | 0.206 | 0.203 | 0.092 | 0.234 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| GTF2F2 |   | 4 | 440 | 957 | 942 | -0.059 | -0.085 | 0.119 | -0.004 | -0.054 |
|---|
| GRIP1 |   | 3 | 557 | 696 | 695 | 0.043 | 0.104 | 0.000 | undef | undef |
|---|
| MSH6 |   | 12 | 140 | 1 | 36 | 0.052 | -0.057 | 0.051 | 0.126 | 0.064 |
|---|
| ZC4H2 |   | 17 | 95 | 570 | 546 | 0.065 | 0.061 | 0.304 | -0.072 | 0.059 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| EDN1 |   | 6 | 301 | 682 | 669 | 0.100 | 0.126 | 0.249 | -0.086 | 0.121 |
|---|
| FHL2 |   | 21 | 73 | 658 | 630 | 0.143 | 0.239 | -0.033 | 0.107 | 0.294 |
|---|
| SKP2 |   | 29 | 45 | 1 | 23 | 0.092 | -0.043 | 0.237 | undef | -0.009 |
|---|
| CD72 |   | 1 | 1195 | 1786 | 1800 | 0.056 | -0.234 | 0.014 | -0.122 | -0.093 |
|---|
| LEPR |   | 2 | 743 | 1461 | 1451 | -0.012 | 0.083 | -0.057 | -0.058 | 0.085 |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| GRB2 |   | 13 | 124 | 318 | 316 | -0.040 | -0.159 | -0.036 | -0.028 | -0.024 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| NAT1 |   | 3 | 557 | 658 | 654 | 0.175 | 0.043 | 0.032 | -0.014 | 0.011 |
|---|
| CEPT1 |   | 2 | 743 | 1510 | 1502 | -0.208 | 0.038 | 0.194 | 0.163 | 0.205 |
|---|
| TFDP1 |   | 6 | 301 | 806 | 783 | 0.123 | -0.178 | 0.129 | undef | -0.065 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| FHL1 |   | 14 | 117 | 614 | 591 | -0.041 | 0.104 | 0.301 | undef | 0.266 |
|---|
| CDK2 |   | 31 | 43 | 179 | 174 | 0.196 | -0.096 | 0.109 | 0.001 | 0.362 |
|---|
| SHROOM2 |   | 3 | 557 | 1396 | 1382 | 0.080 | 0.059 | -0.174 | 0.142 | -0.105 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
GO Enrichment output for subnetwork 6534 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.209E-10 | 2.953E-07 |
|---|
| response to UV | GO:0009411 |  | 5.415E-10 | 6.615E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.283E-10 | 5.931E-07 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 7.604E-10 | 4.644E-07 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.939E-09 | 1.436E-06 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 3.265E-08 | 1.33E-05 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.331E-08 | 1.861E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 5.331E-08 | 1.628E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 7.655E-08 | 2.078E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 7.655E-08 | 1.87E-05 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 9.556E-08 | 2.122E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.285E-10 | 5.498E-07 |
|---|
| response to UV | GO:0009411 |  | 7.5E-10 | 9.022E-07 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 1.236E-09 | 9.913E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.371E-09 | 8.247E-07 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.47E-09 | 2.151E-06 |
|---|
| interphase | GO:0051325 |  | 5.899E-09 | 2.366E-06 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 3.555E-08 | 1.222E-05 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.399E-08 | 2.225E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 7.399E-08 | 1.978E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 8.782E-08 | 2.113E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.147E-07 | 2.509E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.906E-10 | 2.278E-06 |
|---|
| response to UV | GO:0009411 |  | 1.749E-09 | 2.011E-06 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 2.848E-09 | 2.184E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.135E-09 | 2.378E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.706E-08 | 7.846E-06 |
|---|
| interphase | GO:0051325 |  | 2.092E-08 | 8.021E-06 |
|---|
| response to light stimulus | GO:0009416 |  | 4.513E-08 | 1.483E-05 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.068E-07 | 3.071E-05 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.309E-07 | 3.344E-05 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.124E-07 | 4.884E-05 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.124E-07 | 4.44E-05 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| heart development | GO:0007507 |  | 6.086E-09 | 1.122E-05 |
|---|
| negative regulation of blood coagulation | GO:0030195 |  | 1.431E-07 | 1.318E-04 |
|---|
| negative regulation of coagulation | GO:0050819 |  | 4.248E-07 | 2.61E-04 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 6.641E-07 | 3.06E-04 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 9.315E-07 | 3.434E-04 |
|---|
| response to light stimulus | GO:0009416 |  | 1.335E-06 | 4.1E-04 |
|---|
| response to UV | GO:0009411 |  | 1.398E-06 | 3.682E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.424E-06 | 3.281E-04 |
|---|
| negative regulation of multicellular organismal process | GO:0051241 |  | 1.676E-06 | 3.432E-04 |
|---|
| regulation of blood coagulation | GO:0030193 |  | 1.984E-06 | 3.656E-04 |
|---|
| regulation of coagulation | GO:0050818 |  | 3.57E-06 | 5.981E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.906E-10 | 2.278E-06 |
|---|
| response to UV | GO:0009411 |  | 1.749E-09 | 2.011E-06 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 2.848E-09 | 2.184E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.135E-09 | 2.378E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.706E-08 | 7.846E-06 |
|---|
| interphase | GO:0051325 |  | 2.092E-08 | 8.021E-06 |
|---|
| response to light stimulus | GO:0009416 |  | 4.513E-08 | 1.483E-05 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.068E-07 | 3.071E-05 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.309E-07 | 3.344E-05 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.124E-07 | 4.884E-05 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.124E-07 | 4.44E-05 |
|---|



