Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6521
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7553 | 3.428e-03 | 5.540e-04 | 7.670e-01 | 1.457e-06 |
|---|
| Loi | 0.2303 | 8.030e-02 | 1.137e-02 | 4.283e-01 | 3.909e-04 |
|---|
| Schmidt | 0.6727 | 0.000e+00 | 0.000e+00 | 4.572e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.481e-03 | 0.000e+00 |
|---|
| Wang | 0.2613 | 2.714e-03 | 4.415e-02 | 3.979e-01 | 4.767e-05 |
|---|
Expression data for subnetwork 6521 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
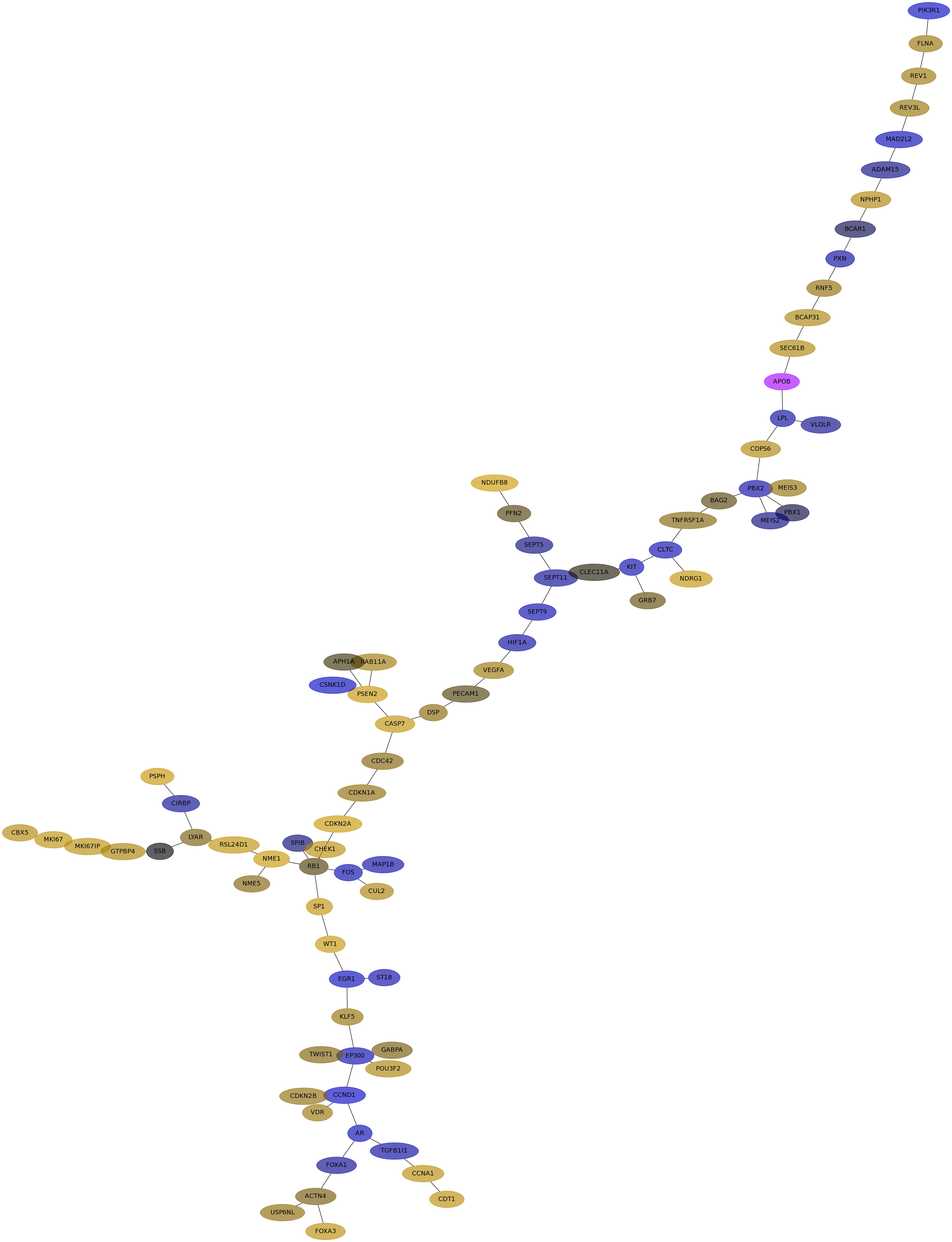
Score for each gene in subnetwork 6521 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| NME5 |   | 1 | 1195 | 1803 | 1818 | 0.046 | 0.054 | 0.044 | 0.131 | 0.038 |
|---|
| NDRG1 |   | 6 | 301 | 658 | 646 | 0.222 | 0.143 | -0.040 | -0.018 | 0.242 |
|---|
| PBX1 |   | 6 | 301 | 1 | 52 | -0.010 | 0.145 | -0.013 | 0.176 | -0.053 |
|---|
| USP6NL |   | 2 | 743 | 1203 | 1206 | 0.060 | -0.111 | undef | undef | undef |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| BCAP31 |   | 2 | 743 | 1803 | 1782 | 0.131 | 0.020 | 0.055 | 0.168 | -0.018 |
|---|
| CCND1 |   | 26 | 52 | 1 | 24 | -0.258 | 0.074 | 0.117 | undef | 0.003 |
|---|
| CDC42 |   | 26 | 52 | 179 | 177 | 0.047 | 0.242 | -0.027 | 0.040 | -0.042 |
|---|
| NDUFB8 |   | 5 | 360 | 696 | 680 | 0.286 | -0.012 | 0.340 | -0.171 | 0.205 |
|---|
| PSEN2 |   | 14 | 117 | 614 | 591 | 0.239 | -0.068 | -0.157 | 0.295 | 0.045 |
|---|
| ST18 |   | 16 | 104 | 236 | 227 | -0.131 | 0.028 | -0.088 | 0.186 | 0.151 |
|---|
| SEPT11 |   | 1 | 1195 | 1803 | 1818 | -0.075 | -0.054 | 0.193 | 0.064 | 0.048 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| MEIS3 |   | 1 | 1195 | 1803 | 1818 | 0.072 | 0.283 | undef | undef | undef |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| MEIS2 |   | 1 | 1195 | 1803 | 1818 | -0.043 | 0.102 | 0.137 | undef | 0.023 |
|---|
| SEC61B |   | 1 | 1195 | 1803 | 1818 | 0.140 | 0.074 | 0.073 | 0.042 | -0.227 |
|---|
| SEPT9 |   | 1 | 1195 | 1803 | 1818 | -0.139 | -0.080 | 0.267 | 0.130 | -0.084 |
|---|
| APH1A |   | 3 | 557 | 1803 | 1777 | 0.009 | -0.165 | 0.107 | 0.183 | -0.098 |
|---|
| GABPA |   | 23 | 66 | 696 | 665 | 0.033 | -0.183 | 0.066 | 0.141 | -0.161 |
|---|
| MKI67 |   | 5 | 360 | 1 | 56 | 0.205 | -0.123 | 0.176 | -0.086 | 0.018 |
|---|
| CIRBP |   | 5 | 360 | 366 | 379 | -0.069 | 0.208 | 0.110 | 0.049 | -0.045 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| DSP |   | 7 | 256 | 590 | 570 | 0.054 | 0.235 | -0.119 | -0.033 | 0.053 |
|---|
| VDR |   | 3 | 557 | 1141 | 1129 | 0.086 | 0.208 | -0.003 | -0.051 | -0.046 |
|---|
| NME1 |   | 1 | 1195 | 1803 | 1818 | 0.267 | 0.106 | 0.046 | -0.059 | -0.035 |
|---|
| CLTC |   | 3 | 557 | 983 | 979 | -0.155 | 0.072 | -0.095 | 0.179 | 0.048 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| TNFRSF1A |   | 8 | 222 | 412 | 413 | 0.050 | 0.094 | -0.014 | 0.214 | -0.045 |
|---|
| ACTN4 |   | 14 | 117 | 318 | 315 | 0.035 | 0.180 | undef | -0.048 | undef |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| LPL |   | 13 | 124 | 236 | 230 | -0.102 | -0.062 | 0.292 | 0.087 | -0.105 |
|---|
| SSB |   | 2 | 743 | 1803 | 1782 | -0.001 | -0.058 | 0.147 | -0.071 | -0.161 |
|---|
| SEPT5 |   | 2 | 743 | 696 | 717 | -0.050 | -0.078 | -0.021 | 0.176 | 0.075 |
|---|
| COPS6 |   | 16 | 104 | 1 | 31 | 0.147 | -0.068 | 0.128 | 0.216 | 0.065 |
|---|
| CSNK1D |   | 6 | 301 | 614 | 602 | -0.222 | -0.044 | 0.170 | -0.088 | 0.013 |
|---|
| PSPH |   | 20 | 81 | 366 | 356 | 0.240 | -0.019 | -0.102 | 0.358 | 0.025 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| CDKN2B |   | 7 | 256 | 236 | 237 | 0.066 | 0.050 | -0.028 | 0.039 | 0.153 |
|---|
| POU3F2 |   | 17 | 95 | 928 | 891 | 0.135 | 0.114 | 0.155 | 0.253 | 0.040 |
|---|
| MKI67IP |   | 2 | 743 | 1803 | 1782 | 0.190 | -0.123 | undef | undef | undef |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| CDT1 |   | 3 | 557 | 1 | 63 | 0.194 | -0.184 | -0.072 | undef | -0.030 |
|---|
| FOXA3 |   | 12 | 140 | 318 | 322 | 0.192 | 0.289 | undef | -0.171 | undef |
|---|
| CHEK1 |   | 18 | 86 | 141 | 136 | 0.189 | -0.134 | 0.119 | -0.088 | -0.054 |
|---|
| SPIB |   | 2 | 743 | 1803 | 1782 | -0.034 | -0.202 | -0.070 | -0.102 | -0.179 |
|---|
| BAG2 |   | 4 | 440 | 1793 | 1755 | 0.016 | 0.178 | 0.152 | -0.249 | 0.083 |
|---|
| PECAM1 |   | 4 | 440 | 83 | 112 | 0.014 | -0.161 | 0.040 | -0.105 | -0.106 |
|---|
| PFN2 |   | 11 | 148 | 141 | 143 | 0.014 | -0.001 | 0.286 | -0.002 | -0.056 |
|---|
| APOB |   | 3 | 557 | 1771 | 1745 | undef | 0.001 | 0.156 | 0.151 | 0.144 |
|---|
| RAB11A |   | 1 | 1195 | 1803 | 1818 | 0.100 | 0.048 | -0.064 | 0.224 | 0.072 |
|---|
| MAD2L2 |   | 2 | 743 | 1803 | 1782 | -0.167 | -0.009 | undef | 0.144 | undef |
|---|
| LYAR |   | 2 | 743 | 1803 | 1782 | 0.037 | -0.154 | undef | -0.245 | undef |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| RSL24D1 |   | 1 | 1195 | 1803 | 1818 | 0.206 | -0.026 | 0.169 | -0.059 | -0.022 |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| ADAM15 |   | 3 | 557 | 1803 | 1777 | -0.048 | -0.078 | -0.208 | 0.172 | -0.048 |
|---|
| REV1 |   | 9 | 196 | 296 | 285 | 0.090 | -0.027 | 0.165 | 0.049 | 0.039 |
|---|
| RNF5 |   | 2 | 743 | 1102 | 1102 | 0.067 | 0.024 | 0.050 | 0.035 | 0.252 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| PXN |   | 12 | 140 | 1 | 36 | -0.088 | 0.284 | -0.022 | 0.060 | 0.114 |
|---|
| CLEC11A |   | 7 | 256 | 692 | 673 | 0.004 | 0.353 | 0.005 | 0.081 | 0.272 |
|---|
| VLDLR |   | 1 | 1195 | 1803 | 1818 | -0.059 | 0.109 | -0.128 | 0.007 | 0.024 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| GTPBP4 |   | 4 | 440 | 882 | 870 | 0.129 | -0.179 | 0.076 | -0.043 | -0.026 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| BCAR1 |   | 6 | 301 | 1 | 52 | -0.012 | 0.229 | undef | 0.223 | undef |
|---|
| PBX2 |   | 5 | 360 | 1 | 56 | -0.122 | -0.073 | 0.264 | -0.095 | 0.092 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| REV3L |   | 2 | 743 | 806 | 806 | 0.080 | 0.041 | 0.146 | -0.076 | 0.257 |
|---|
| CASP7 |   | 6 | 301 | 614 | 602 | 0.216 | 0.095 | 0.255 | undef | 0.035 |
|---|
| HIF1A |   | 13 | 124 | 236 | 230 | -0.098 | 0.224 | -0.118 | 0.135 | 0.356 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| CUL2 |   | 7 | 256 | 842 | 816 | 0.124 | -0.192 | -0.090 | -0.035 | 0.166 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| CBX5 |   | 5 | 360 | 1 | 56 | 0.155 | 0.016 | 0.148 | 0.030 | -0.009 |
|---|
| NPHP1 |   | 1 | 1195 | 1803 | 1818 | 0.139 | 0.095 | 0.026 | -0.235 | -0.110 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| FOXA1 |   | 27 | 50 | 179 | 176 | -0.072 | 0.071 | -0.015 | 0.214 | 0.005 |
|---|
GO Enrichment output for subnetwork 6521 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.327E-10 | 2.279E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.864E-09 | 4.72E-06 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 5.624E-09 | 4.58E-06 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 6.044E-08 | 3.691E-05 |
|---|
| UTP metabolic process | GO:0046051 |  | 1.083E-07 | 5.293E-05 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.083E-07 | 4.411E-05 |
|---|
| CTP metabolic process | GO:0046036 |  | 1.798E-07 | 6.274E-05 |
|---|
| GTP metabolic process | GO:0046039 |  | 1.798E-07 | 5.49E-05 |
|---|
| sex determination | GO:0007530 |  | 1.843E-07 | 5.004E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.97E-07 | 4.812E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.294E-07 | 5.095E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.792E-09 | 4.313E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.015E-09 | 7.236E-06 |
|---|
| interphase | GO:0051325 |  | 7.933E-09 | 6.362E-06 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 8.534E-09 | 5.133E-06 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 4.259E-08 | 2.049E-05 |
|---|
| macrophage differentiation | GO:0030225 |  | 4.259E-08 | 1.708E-05 |
|---|
| UTP metabolic process | GO:0046051 |  | 8.478E-08 | 2.914E-05 |
|---|
| CTP metabolic process | GO:0046036 |  | 1.519E-07 | 4.568E-05 |
|---|
| GTP metabolic process | GO:0046039 |  | 1.519E-07 | 4.061E-05 |
|---|
| sex determination | GO:0007530 |  | 2.224E-07 | 5.351E-05 |
|---|
| pyrimidine nucleoside triphosphate biosynthetic process | GO:0009148 |  | 2.52E-07 | 5.512E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| macrophage differentiation | GO:0030225 |  | 3.35E-08 | 7.705E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.083E-08 | 6.996E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 7.774E-08 | 5.96E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.503E-07 | 8.643E-05 |
|---|
| UTP metabolic process | GO:0046051 |  | 1.547E-07 | 7.114E-05 |
|---|
| sex determination | GO:0007530 |  | 1.639E-07 | 6.283E-05 |
|---|
| interphase | GO:0051325 |  | 1.8E-07 | 5.914E-05 |
|---|
| CTP metabolic process | GO:0046036 |  | 2.769E-07 | 7.96E-05 |
|---|
| GTP metabolic process | GO:0046039 |  | 2.769E-07 | 7.076E-05 |
|---|
| pyrimidine nucleoside triphosphate biosynthetic process | GO:0009148 |  | 4.59E-07 | 1.056E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.038E-07 | 1.053E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| prostate gland development | GO:0030850 |  | 7.578E-06 | 0.01396577 |
|---|
| CTP metabolic process | GO:0046036 |  | 7.578E-06 | 6.983E-03 |
|---|
| GTP metabolic process | GO:0046039 |  | 7.578E-06 | 4.655E-03 |
|---|
| amyloid precursor protein catabolic process | GO:0042987 |  | 7.578E-06 | 3.491E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.505E-05 | 5.549E-03 |
|---|
| pyrimidine nucleoside triphosphate biosynthetic process | GO:0009148 |  | 1.505E-05 | 4.624E-03 |
|---|
| cellular response to oxidative stress | GO:0034599 |  | 2.377E-05 | 6.259E-03 |
|---|
| amyloid precursor protein metabolic process | GO:0042982 |  | 2.617E-05 | 6.029E-03 |
|---|
| membrane protein ectodomain proteolysis | GO:0006509 |  | 2.617E-05 | 5.359E-03 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 2.682E-05 | 4.943E-03 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 3.442E-05 | 5.766E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| macrophage differentiation | GO:0030225 |  | 3.35E-08 | 7.705E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.083E-08 | 6.996E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 7.774E-08 | 5.96E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.503E-07 | 8.643E-05 |
|---|
| UTP metabolic process | GO:0046051 |  | 1.547E-07 | 7.114E-05 |
|---|
| sex determination | GO:0007530 |  | 1.639E-07 | 6.283E-05 |
|---|
| interphase | GO:0051325 |  | 1.8E-07 | 5.914E-05 |
|---|
| CTP metabolic process | GO:0046036 |  | 2.769E-07 | 7.96E-05 |
|---|
| GTP metabolic process | GO:0046039 |  | 2.769E-07 | 7.076E-05 |
|---|
| pyrimidine nucleoside triphosphate biosynthetic process | GO:0009148 |  | 4.59E-07 | 1.056E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.038E-07 | 1.053E-04 |
|---|



