Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6515
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7552 | 3.431e-03 | 5.540e-04 | 7.672e-01 | 1.458e-06 |
|---|
| Loi | 0.2302 | 8.037e-02 | 1.139e-02 | 4.285e-01 | 3.922e-04 |
|---|
| Schmidt | 0.6725 | 0.000e+00 | 0.000e+00 | 4.590e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.472e-03 | 0.000e+00 |
|---|
| Wang | 0.2613 | 2.720e-03 | 4.420e-02 | 3.981e-01 | 4.787e-05 |
|---|
Expression data for subnetwork 6515 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
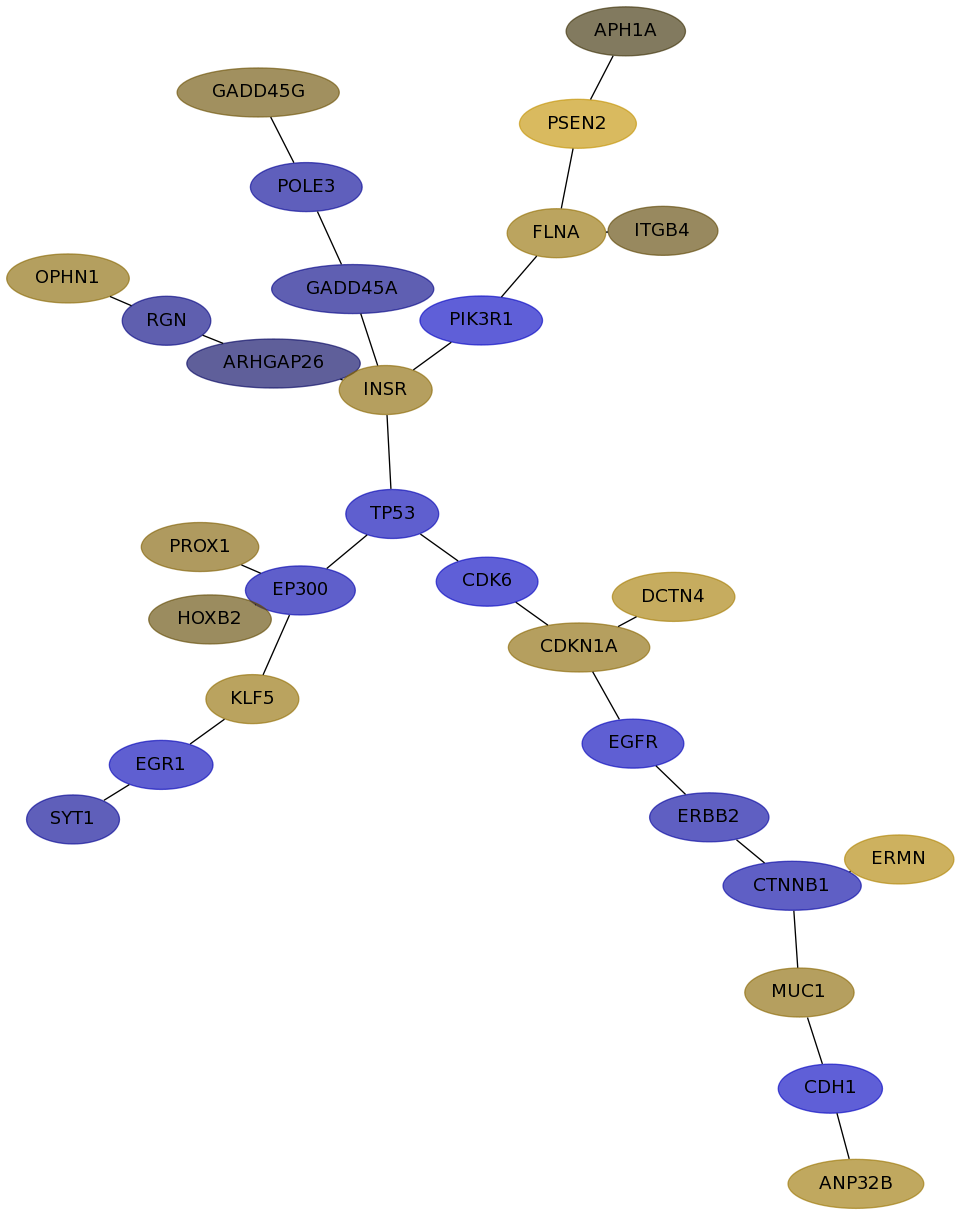
Score for each gene in subnetwork 6515 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| ARHGAP26 |   | 21 | 73 | 457 | 438 | -0.023 | 0.171 | -0.222 | 0.015 | -0.348 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| APH1A |   | 3 | 557 | 1803 | 1777 | 0.009 | -0.165 | 0.107 | 0.183 | -0.098 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| OPHN1 |   | 2 | 743 | 1822 | 1806 | 0.062 | 0.278 | 0.194 | 0.008 | -0.182 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| PROX1 |   | 4 | 440 | 614 | 618 | 0.050 | -0.030 | -0.128 | 0.161 | 0.112 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| SYT1 |   | 11 | 148 | 1 | 39 | -0.079 | 0.051 | 0.038 | 0.189 | -0.124 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| RGN |   | 6 | 301 | 570 | 557 | -0.051 | -0.050 | 0.184 | 0.139 | -0.120 |
|---|
| MUC1 |   | 13 | 124 | 1547 | 1490 | 0.063 | 0.214 | -0.190 | undef | -0.132 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| GADD45A |   | 11 | 148 | 1351 | 1308 | -0.057 | 0.076 | -0.099 | 0.109 | 0.093 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| DCTN4 |   | 5 | 360 | 1010 | 987 | 0.122 | -0.025 | -0.111 | 0.173 | -0.105 |
|---|
| PSEN2 |   | 14 | 117 | 614 | 591 | 0.239 | -0.068 | -0.157 | 0.295 | 0.045 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| ANP32B |   | 2 | 743 | 1822 | 1806 | 0.096 | 0.080 | 0.309 | -0.081 | 0.045 |
|---|
| POLE3 |   | 17 | 95 | 141 | 138 | -0.083 | -0.033 | 0.231 | 0.096 | 0.011 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| ERMN |   | 1 | 1195 | 1822 | 1837 | 0.157 | 0.068 | undef | -0.061 | undef |
|---|
| CDK6 |   | 16 | 104 | 318 | 314 | -0.229 | 0.107 | -0.028 | -0.118 | 0.104 |
|---|
GO Enrichment output for subnetwork 6515 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Notch receptor processing | GO:0007220 |  | 1.094E-07 | 2.673E-04 |
|---|
| amyloid precursor protein catabolic process | GO:0042987 |  | 3.818E-07 | 4.663E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 4.674E-07 | 3.806E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 6.098E-07 | 3.725E-04 |
|---|
| amyloid precursor protein metabolic process | GO:0042982 |  | 9.133E-07 | 4.462E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 1.006E-06 | 4.098E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.788E-06 | 6.24E-04 |
|---|
| positive regulation of transcription factor import into nucleus | GO:0042993 |  | 2.38E-06 | 7.269E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.933E-06 | 7.962E-04 |
|---|
| membrane protein proteolysis | GO:0033619 |  | 3.089E-06 | 7.547E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 3.304E-06 | 7.338E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Notch receptor processing | GO:0007220 |  | 1.478E-07 | 3.556E-04 |
|---|
| amyloid precursor protein catabolic process | GO:0042987 |  | 5.154E-07 | 6.201E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 5.96E-07 | 4.78E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 8.232E-07 | 4.952E-04 |
|---|
| amyloid precursor protein metabolic process | GO:0042982 |  | 1.233E-06 | 5.931E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 1.532E-06 | 6.145E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.758E-06 | 6.042E-04 |
|---|
| positive regulation of transcription factor import into nucleus | GO:0042993 |  | 3.211E-06 | 9.657E-04 |
|---|
| membrane protein proteolysis | GO:0033619 |  | 3.211E-06 | 8.584E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 4.066E-06 | 9.783E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.07E-06 | 8.902E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 4.52E-07 | 1.04E-03 |
|---|
| amyloid precursor protein catabolic process | GO:0042987 |  | 4.649E-07 | 5.347E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 1.251E-06 | 9.589E-04 |
|---|
| amyloid precursor protein metabolic process | GO:0042982 |  | 1.296E-06 | 7.454E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.296E-06 | 5.964E-04 |
|---|
| positive regulation of transcription factor import into nucleus | GO:0042993 |  | 2.767E-06 | 1.061E-03 |
|---|
| membrane protein proteolysis | GO:0033619 |  | 3.796E-06 | 1.247E-03 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 4.9E-06 | 1.409E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.036E-06 | 1.287E-03 |
|---|
| glycoprotein catabolic process | GO:0006516 |  | 5.051E-06 | 1.162E-03 |
|---|
| interphase | GO:0051325 |  | 5.64E-06 | 1.179E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| amyloid precursor protein catabolic process | GO:0042987 |  | 2.827E-07 | 5.21E-04 |
|---|
| positive regulation of transcription factor import into nucleus | GO:0042993 |  | 5.642E-07 | 5.199E-04 |
|---|
| amyloid precursor protein metabolic process | GO:0042982 |  | 9.853E-07 | 6.053E-04 |
|---|
| membrane protein ectodomain proteolysis | GO:0006509 |  | 9.853E-07 | 4.54E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.527E-06 | 5.628E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.768E-06 | 5.429E-04 |
|---|
| positive regulation of protein import into nucleus | GO:0042307 |  | 2.355E-06 | 6.2E-04 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 4.607E-06 | 1.061E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 4.607E-06 | 9.433E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 5.994E-06 | 1.105E-03 |
|---|
| positive regulation of protein kinase activity | GO:0045860 |  | 6.047E-06 | 1.013E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 4.52E-07 | 1.04E-03 |
|---|
| amyloid precursor protein catabolic process | GO:0042987 |  | 4.649E-07 | 5.347E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 1.251E-06 | 9.589E-04 |
|---|
| amyloid precursor protein metabolic process | GO:0042982 |  | 1.296E-06 | 7.454E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.296E-06 | 5.964E-04 |
|---|
| positive regulation of transcription factor import into nucleus | GO:0042993 |  | 2.767E-06 | 1.061E-03 |
|---|
| membrane protein proteolysis | GO:0033619 |  | 3.796E-06 | 1.247E-03 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 4.9E-06 | 1.409E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.036E-06 | 1.287E-03 |
|---|
| glycoprotein catabolic process | GO:0006516 |  | 5.051E-06 | 1.162E-03 |
|---|
| interphase | GO:0051325 |  | 5.64E-06 | 1.179E-03 |
|---|



