Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6485
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7551 | 3.436e-03 | 5.550e-04 | 7.674e-01 | 1.463e-06 |
|---|
| Loi | 0.2302 | 8.043e-02 | 1.140e-02 | 4.286e-01 | 3.930e-04 |
|---|
| Schmidt | 0.6723 | 0.000e+00 | 0.000e+00 | 4.607e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.464e-03 | 0.000e+00 |
|---|
| Wang | 0.2611 | 2.736e-03 | 4.435e-02 | 3.988e-01 | 4.839e-05 |
|---|
Expression data for subnetwork 6485 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
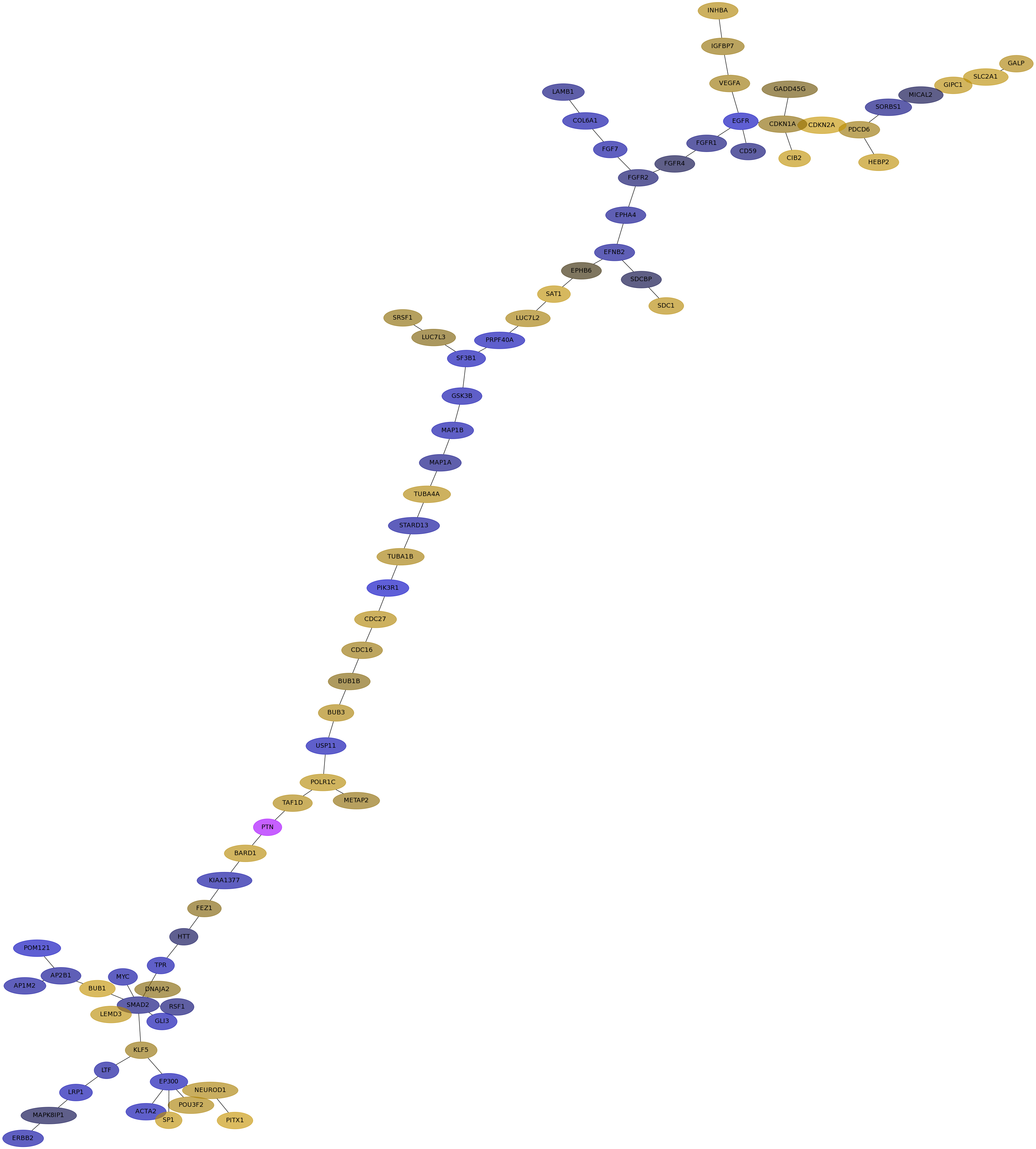
Score for each gene in subnetwork 6485 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| SF3B1 |   | 18 | 86 | 296 | 280 | -0.154 | 0.126 | -0.073 | -0.131 | -0.204 |
|---|
| EPHA4 |   | 1 | 1195 | 1954 | 1959 | -0.061 | -0.095 | -0.008 | 0.042 | -0.113 |
|---|
| SLC2A1 |   | 7 | 256 | 236 | 237 | 0.205 | 0.143 | -0.050 | -0.023 | 0.086 |
|---|
| CIB2 |   | 18 | 86 | 590 | 564 | 0.228 | -0.149 | 0.106 | 0.127 | 0.190 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| GIPC1 |   | 4 | 440 | 1396 | 1379 | 0.185 | 0.124 | -0.045 | -0.095 | 0.006 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| SDC1 |   | 2 | 743 | 1954 | 1933 | 0.172 | 0.241 | -0.088 | 0.179 | 0.059 |
|---|
| LTF |   | 3 | 557 | 1222 | 1210 | -0.082 | 0.060 | -0.268 | -0.081 | -0.280 |
|---|
| IGFBP7 |   | 1 | 1195 | 1954 | 1959 | 0.079 | 0.072 | 0.141 | 0.014 | 0.151 |
|---|
| AP1M2 |   | 1 | 1195 | 1954 | 1959 | -0.082 | 0.154 | -0.068 | 0.005 | 0.056 |
|---|
| AP2B1 |   | 6 | 301 | 780 | 764 | -0.070 | -0.106 | 0.070 | 0.252 | 0.094 |
|---|
| EPHB6 |   | 1 | 1195 | 1954 | 1959 | 0.008 | 0.042 | -0.219 | 0.152 | -0.059 |
|---|
| TUBA4A |   | 7 | 256 | 1 | 47 | 0.157 | 0.136 | -0.010 | undef | 0.156 |
|---|
| METAP2 |   | 2 | 743 | 1294 | 1286 | 0.067 | 0.050 | 0.177 | undef | -0.085 |
|---|
| SAT1 |   | 6 | 301 | 236 | 239 | 0.219 | 0.010 | -0.066 | -0.119 | 0.063 |
|---|
| LRP1 |   | 2 | 743 | 1954 | 1933 | -0.138 | 0.026 | 0.119 | 0.063 | 0.112 |
|---|
| FEZ1 |   | 8 | 222 | 141 | 149 | 0.047 | -0.020 | 0.154 | 0.165 | -0.128 |
|---|
| BUB1B |   | 4 | 440 | 1010 | 991 | 0.050 | -0.164 | 0.037 | -0.101 | 0.148 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| FGFR1 |   | 26 | 52 | 638 | 615 | -0.036 | 0.200 | 0.087 | undef | -0.015 |
|---|
| SRSF1 |   | 1 | 1195 | 1954 | 1959 | 0.065 | -0.077 | 0.127 | undef | -0.085 |
|---|
| TUBA1B |   | 6 | 301 | 590 | 582 | 0.116 | 0.094 | 0.249 | 0.101 | 0.128 |
|---|
| CDC16 |   | 2 | 743 | 1010 | 1007 | 0.086 | -0.029 | 0.095 | 0.029 | 0.022 |
|---|
| SMAD2 |   | 4 | 440 | 590 | 585 | -0.039 | -0.199 | 0.076 | -0.003 | 0.247 |
|---|
| LUC7L3 |   | 16 | 104 | 296 | 281 | 0.043 | 0.237 | 0.175 | 0.135 | 0.074 |
|---|
| LAMB1 |   | 3 | 557 | 1077 | 1068 | -0.041 | 0.226 | -0.020 | -0.011 | 0.238 |
|---|
| FGF7 |   | 10 | 167 | 780 | 760 | -0.094 | 0.237 | 0.070 | 0.038 | 0.083 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| USP11 |   | 7 | 256 | 478 | 462 | -0.144 | 0.106 | 0.121 | 0.218 | -0.044 |
|---|
| STARD13 |   | 2 | 743 | 1879 | 1866 | -0.083 | 0.006 | 0.126 | 0.290 | 0.028 |
|---|
| PTN |   | 24 | 62 | 236 | 225 | undef | 0.022 | 0.000 | 0.054 | -0.092 |
|---|
| EFNB2 |   | 1 | 1195 | 1954 | 1959 | -0.070 | 0.156 | 0.193 | -0.135 | -0.116 |
|---|
| POU3F2 |   | 17 | 95 | 928 | 891 | 0.135 | 0.114 | 0.155 | 0.253 | 0.040 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| RSF1 |   | 2 | 743 | 780 | 787 | -0.032 | 0.063 | 0.103 | 0.218 | 0.021 |
|---|
| KIAA1377 |   | 18 | 86 | 141 | 136 | -0.095 | 0.116 | undef | 0.198 | undef |
|---|
| POM121 |   | 7 | 256 | 780 | 763 | -0.197 | 0.101 | 0.079 | undef | 0.017 |
|---|
| CD59 |   | 1 | 1195 | 1954 | 1959 | -0.032 | 0.228 | 0.013 | -0.039 | -0.010 |
|---|
| GLI3 |   | 21 | 73 | 478 | 456 | -0.138 | 0.080 | -0.007 | 0.282 | -0.018 |
|---|
| MAP1A |   | 12 | 140 | 1 | 36 | -0.045 | 0.143 | -0.054 | 0.162 | 0.147 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| COL6A1 |   | 5 | 360 | 780 | 767 | -0.112 | 0.193 | -0.013 | undef | 0.039 |
|---|
| ACTA2 |   | 2 | 743 | 1949 | 1932 | -0.147 | 0.256 | 0.125 | 0.102 | 0.220 |
|---|
| BUB1 |   | 6 | 301 | 590 | 582 | 0.246 | -0.172 | 0.013 | -0.007 | 0.170 |
|---|
| SDCBP |   | 3 | 557 | 1433 | 1415 | -0.010 | -0.059 | -0.141 | 0.279 | 0.131 |
|---|
| LUC7L2 |   | 1 | 1195 | 1954 | 1959 | 0.116 | -0.120 | 0.065 | -0.168 | 0.107 |
|---|
| INHBA |   | 2 | 743 | 1752 | 1744 | 0.151 | 0.188 | 0.037 | 0.150 | 0.143 |
|---|
| PRPF40A |   | 2 | 743 | 478 | 490 | -0.153 | -0.047 | -0.133 | undef | 0.155 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| FGFR4 |   | 21 | 73 | 179 | 183 | -0.011 | 0.188 | -0.089 | -0.018 | -0.049 |
|---|
| MICAL2 |   | 4 | 440 | 1659 | 1629 | -0.012 | 0.143 | -0.042 | -0.145 | 0.161 |
|---|
| BUB3 |   | 5 | 360 | 478 | 476 | 0.133 | -0.053 | 0.165 | 0.195 | 0.181 |
|---|
| TAF1D |   | 10 | 167 | 590 | 566 | 0.141 | 0.113 | 0.041 | undef | 0.059 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| NEUROD1 |   | 1 | 1195 | 1954 | 1959 | 0.135 | 0.022 | 0.000 | 0.044 | undef |
|---|
| HEBP2 |   | 2 | 743 | 83 | 123 | 0.207 | -0.044 | -0.153 | -0.038 | 0.013 |
|---|
| MAPK8IP1 |   | 4 | 440 | 1576 | 1543 | -0.012 | 0.058 | -0.020 | 0.282 | -0.049 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| GALP |   | 1 | 1195 | 1954 | 1959 | 0.136 | -0.069 | undef | undef | undef |
|---|
| TPR |   | 11 | 148 | 727 | 701 | -0.130 | 0.120 | -0.036 | -0.085 | 0.156 |
|---|
| PDCD6 |   | 10 | 167 | 1 | 40 | 0.091 | -0.060 | undef | undef | undef |
|---|
| CDC27 |   | 6 | 301 | 1010 | 986 | 0.156 | -0.086 | -0.029 | 0.006 | 0.348 |
|---|
| HTT |   | 14 | 117 | 141 | 140 | -0.016 | -0.015 | 0.075 | 0.092 | -0.093 |
|---|
| FGFR2 |   | 7 | 256 | 179 | 201 | -0.024 | -0.001 | 0.129 | undef | 0.009 |
|---|
| LEMD3 |   | 1 | 1195 | 1954 | 1959 | 0.196 | 0.154 | 0.081 | 0.082 | 0.236 |
|---|
| PITX1 |   | 3 | 557 | 236 | 265 | 0.256 | 0.046 | 0.091 | -0.068 | 0.139 |
|---|
| POLR1C |   | 3 | 557 | 1323 | 1309 | 0.176 | -0.210 | 0.181 | -0.126 | 0.038 |
|---|
| DNAJA2 |   | 3 | 557 | 780 | 781 | 0.054 | 0.063 | -0.152 | -0.019 | 0.081 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
GO Enrichment output for subnetwork 6485 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.098E-08 | 5.125E-05 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 3.96E-06 | 4.838E-03 |
|---|
| negative regulation of B cell activation | GO:0050869 |  | 4.49E-06 | 3.656E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 7.821E-06 | 4.777E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.258E-06 | 4.524E-03 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 9.509E-06 | 3.872E-03 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 1.246E-05 | 4.348E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.246E-05 | 3.804E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.328E-05 | 3.606E-03 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.328E-05 | 3.245E-03 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.356E-05 | 3.011E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 5.574E-07 | 1.341E-03 |
|---|
| negative regulation of B cell activation | GO:0050869 |  | 5.517E-06 | 6.637E-03 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 5.837E-06 | 4.681E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 9.607E-06 | 5.779E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.208E-05 | 5.814E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.286E-05 | 5.156E-03 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.396E-05 | 4.799E-03 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 1.53E-05 | 4.601E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.53E-05 | 4.09E-03 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.737E-05 | 4.18E-03 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.987E-05 | 4.346E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.73E-06 | 3.98E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.264E-05 | 0.01454077 |
|---|
| negative regulation of B cell activation | GO:0050869 |  | 1.264E-05 | 9.694E-03 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 1.465E-05 | 8.423E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.923E-05 | 8.847E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.43E-05 | 0.01315014 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.495E-05 | 0.01148455 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 3.495E-05 | 0.01004898 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 3.495E-05 | 8.932E-03 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 3.518E-05 | 8.092E-03 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 4.277E-05 | 8.942E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 2.968E-06 | 5.47E-03 |
|---|
| lung development | GO:0030324 |  | 8.092E-06 | 7.457E-03 |
|---|
| respiratory tube development | GO:0030323 |  | 1.056E-05 | 6.49E-03 |
|---|
| respiratory system development | GO:0060541 |  | 1.056E-05 | 4.868E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 3.51E-05 | 0.01293806 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 3.523E-05 | 0.01082005 |
|---|
| anterior/posterior pattern formation | GO:0009952 |  | 3.646E-05 | 9.6E-03 |
|---|
| cell fate commitment | GO:0045165 |  | 4.421E-05 | 0.01018501 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 4.832E-05 | 9.894E-03 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 6.465E-05 | 0.01191427 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 6.82E-05 | 0.0114264 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.73E-06 | 3.98E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.264E-05 | 0.01454077 |
|---|
| negative regulation of B cell activation | GO:0050869 |  | 1.264E-05 | 9.694E-03 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 1.465E-05 | 8.423E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.923E-05 | 8.847E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.43E-05 | 0.01315014 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.495E-05 | 0.01148455 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 3.495E-05 | 0.01004898 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 3.495E-05 | 8.932E-03 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 3.518E-05 | 8.092E-03 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 4.277E-05 | 8.942E-03 |
|---|



