Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6466
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7557 | 3.403e-03 | 5.490e-04 | 7.660e-01 | 1.431e-06 |
|---|
| Loi | 0.2298 | 8.106e-02 | 1.157e-02 | 4.301e-01 | 4.034e-04 |
|---|
| Schmidt | 0.6726 | 0.000e+00 | 0.000e+00 | 4.578e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.476e-03 | 0.000e+00 |
|---|
| Wang | 0.2611 | 2.747e-03 | 4.444e-02 | 3.993e-01 | 4.875e-05 |
|---|
Expression data for subnetwork 6466 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
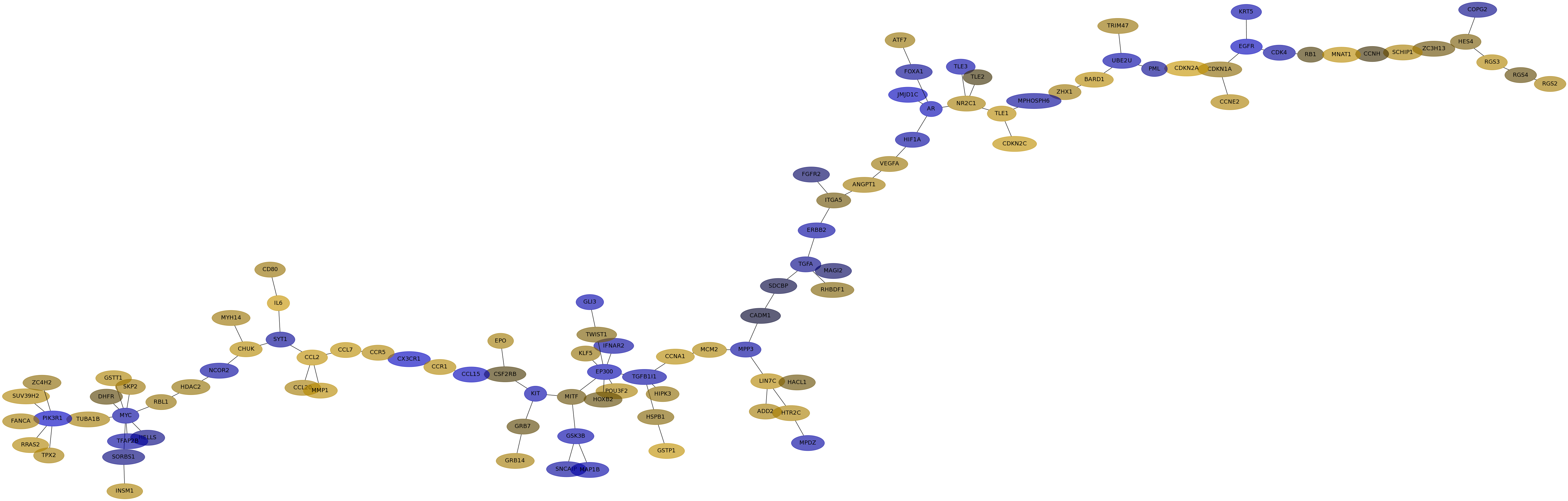
Score for each gene in subnetwork 6466 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| RGS4 |   | 4 | 440 | 842 | 831 | 0.022 | 0.160 | -0.006 | undef | 0.371 |
|---|
| CCNE2 |   | 11 | 148 | 412 | 407 | 0.136 | -0.178 | 0.241 | undef | -0.055 |
|---|
| IFNAR2 |   | 4 | 440 | 1558 | 1528 | -0.100 | 0.055 | 0.106 | -0.294 | -0.058 |
|---|
| HACL1 |   | 4 | 440 | 590 | 585 | 0.028 | -0.071 | undef | -0.278 | undef |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| CCL2 |   | 1 | 1195 | 1846 | 1856 | 0.212 | -0.165 | 0.130 | -0.032 | -0.207 |
|---|
| TPX2 |   | 5 | 360 | 412 | 416 | 0.114 | -0.121 | -0.018 | undef | -0.056 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| ADD2 |   | 8 | 222 | 614 | 599 | 0.113 | -0.022 | -0.061 | 0.270 | 0.180 |
|---|
| CHUK |   | 6 | 301 | 318 | 330 | 0.173 | 0.080 | 0.152 | undef | 0.215 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| ZC3H13 |   | 1 | 1195 | 1846 | 1856 | 0.029 | -0.086 | 0.015 | -0.062 | 0.058 |
|---|
| CDKN2C |   | 16 | 104 | 236 | 227 | 0.216 | -0.106 | 0.202 | -0.024 | -0.081 |
|---|
| RGS3 |   | 2 | 743 | 1846 | 1832 | 0.153 | 0.080 | 0.128 | undef | 0.037 |
|---|
| MITF |   | 5 | 360 | 696 | 680 | 0.027 | 0.032 | 0.065 | 0.145 | 0.183 |
|---|
| RHBDF1 |   | 1 | 1195 | 1846 | 1856 | 0.048 | 0.237 | -0.142 | undef | -0.015 |
|---|
| UBE2U |   | 2 | 743 | 1846 | 1832 | -0.113 | 0.255 | undef | undef | undef |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| MYH14 |   | 2 | 743 | 1786 | 1776 | 0.092 | 0.103 | -0.105 | undef | -0.118 |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| NR2C1 |   | 8 | 222 | 727 | 703 | 0.121 | 0.039 | 0.119 | -0.197 | -0.026 |
|---|
| KRT5 |   | 10 | 167 | 806 | 780 | -0.125 | 0.046 | 0.064 | -0.006 | -0.190 |
|---|
| CCNH |   | 2 | 743 | 570 | 593 | 0.010 | -0.066 | 0.071 | -0.042 | 0.049 |
|---|
| CCL15 |   | 2 | 743 | 412 | 439 | -0.190 | 0.004 | 0.141 | -0.215 | 0.007 |
|---|
| TFAP2B |   | 1 | 1195 | 1846 | 1856 | -0.101 | 0.107 | -0.059 | 0.102 | -0.086 |
|---|
| CDK4 |   | 4 | 440 | 913 | 904 | -0.092 | -0.201 | 0.091 | -0.043 | -0.017 |
|---|
| TGFA |   | 3 | 557 | 1396 | 1382 | -0.057 | -0.027 | -0.158 | -0.216 | 0.020 |
|---|
| MMP1 |   | 11 | 148 | 366 | 360 | 0.192 | 0.014 | 0.036 | 0.044 | 0.260 |
|---|
| CADM1 |   | 1 | 1195 | 1846 | 1856 | -0.006 | 0.075 | 0.141 | -0.078 | 0.072 |
|---|
| MCM2 |   | 2 | 743 | 1846 | 1832 | 0.148 | -0.200 | 0.074 | -0.045 | 0.168 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| TUBA1B |   | 6 | 301 | 590 | 582 | 0.116 | 0.094 | 0.249 | 0.101 | 0.128 |
|---|
| TRIM47 |   | 1 | 1195 | 1846 | 1856 | 0.083 | 0.109 | undef | undef | undef |
|---|
| HELLS |   | 2 | 743 | 1846 | 1832 | -0.050 | -0.018 | 0.199 | 0.057 | 0.107 |
|---|
| RBL1 |   | 21 | 73 | 1 | 27 | 0.078 | undef | undef | 0.051 | undef |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| TLE1 |   | 4 | 440 | 727 | 723 | 0.183 | 0.050 | 0.030 | 0.080 | 0.055 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| MPP3 |   | 4 | 440 | 1650 | 1612 | -0.095 | -0.007 | -0.098 | 0.087 | -0.046 |
|---|
| MAGI2 |   | 1 | 1195 | 1846 | 1856 | -0.022 | 0.024 | -0.020 | 0.134 | 0.211 |
|---|
| HSPB1 |   | 22 | 69 | 1 | 25 | 0.053 | 0.169 | 0.017 | 0.179 | 0.161 |
|---|
| ANGPT1 |   | 5 | 360 | 696 | 680 | 0.106 | -0.167 | 0.084 | -0.035 | 0.250 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| HDAC2 |   | 13 | 124 | 318 | 316 | 0.079 | -0.094 | 0.148 | 0.089 | 0.037 |
|---|
| POU3F2 |   | 17 | 95 | 928 | 891 | 0.135 | 0.114 | 0.155 | 0.253 | 0.040 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| GRB14 |   | 24 | 62 | 179 | 181 | 0.113 | -0.129 | 0.143 | -0.098 | 0.138 |
|---|
| HTR2C |   | 2 | 743 | 842 | 854 | 0.135 | 0.027 | 0.162 | 0.138 | undef |
|---|
| CX3CR1 |   | 2 | 743 | 412 | 439 | -0.213 | 0.060 | 0.113 | -0.040 | -0.050 |
|---|
| ATF7 |   | 28 | 47 | 179 | 175 | 0.085 | 0.115 | -0.088 | 0.172 | -0.092 |
|---|
| GSTT1 |   | 15 | 111 | 1 | 32 | 0.148 | 0.107 | -0.190 | 0.022 | -0.064 |
|---|
| RGS2 |   | 3 | 557 | 1131 | 1126 | 0.118 | 0.061 | 0.084 | 0.050 | 0.178 |
|---|
| GSTP1 |   | 14 | 117 | 1 | 34 | 0.222 | -0.166 | 0.201 | 0.126 | -0.136 |
|---|
| COPG2 |   | 4 | 440 | 412 | 419 | -0.058 | -0.232 | 0.000 | 0.086 | -0.081 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| CSF2RB |   | 3 | 557 | 1492 | 1464 | 0.013 | -0.196 | -0.109 | 0.012 | -0.141 |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| GLI3 |   | 21 | 73 | 478 | 456 | -0.138 | 0.080 | -0.007 | 0.282 | -0.018 |
|---|
| SUV39H2 |   | 8 | 222 | 318 | 325 | 0.150 | -0.097 | -0.057 | undef | 0.028 |
|---|
| CCR1 |   | 2 | 743 | 412 | 439 | 0.162 | -0.205 | -0.049 | -0.123 | -0.109 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| SCHIP1 |   | 10 | 167 | 478 | 459 | 0.130 | 0.136 | 0.105 | -0.022 | 0.198 |
|---|
| INSM1 |   | 23 | 66 | 570 | 545 | 0.135 | -0.184 | -0.188 | 0.237 | -0.032 |
|---|
| MPDZ |   | 3 | 557 | 1846 | 1817 | -0.110 | 0.062 | 0.231 | 0.291 | 0.167 |
|---|
| SDCBP |   | 3 | 557 | 1433 | 1415 | -0.010 | -0.059 | -0.141 | 0.279 | 0.131 |
|---|
| RRAS2 |   | 7 | 256 | 1681 | 1633 | 0.155 | 0.076 | 0.289 | 0.044 | 0.031 |
|---|
| MPHOSPH6 |   | 2 | 743 | 1846 | 1832 | -0.084 | 0.175 | -0.269 | 0.082 | 0.146 |
|---|
| TLE2 |   | 4 | 440 | 806 | 790 | 0.010 | 0.052 | 0.202 | undef | 0.093 |
|---|
| CCL7 |   | 2 | 743 | 412 | 439 | 0.204 | -0.142 | 0.011 | undef | -0.140 |
|---|
| HES4 |   | 1 | 1195 | 1846 | 1856 | 0.040 | 0.192 | undef | -0.114 | undef |
|---|
| CCL20 |   | 1 | 1195 | 1846 | 1856 | 0.118 | 0.001 | -0.032 | -0.133 | -0.165 |
|---|
| TLE3 |   | 1 | 1195 | 1846 | 1856 | -0.121 | 0.084 | 0.058 | 0.100 | -0.097 |
|---|
| ZC4H2 |   | 17 | 95 | 570 | 546 | 0.065 | 0.061 | 0.304 | -0.072 | 0.059 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| JMJD1C |   | 7 | 256 | 552 | 542 | -0.195 | 0.241 | 0.074 | undef | 0.129 |
|---|
| SKP2 |   | 29 | 45 | 1 | 23 | 0.092 | -0.043 | 0.237 | undef | -0.009 |
|---|
| ITGA5 |   | 26 | 52 | 179 | 177 | 0.031 | 0.192 | 0.125 | 0.133 | 0.202 |
|---|
| CD80 |   | 4 | 440 | 1055 | 1035 | 0.081 | -0.199 | -0.136 | 0.034 | -0.078 |
|---|
| SYT1 |   | 11 | 148 | 1 | 39 | -0.079 | 0.051 | 0.038 | 0.189 | -0.124 |
|---|
| LIN7C |   | 10 | 167 | 590 | 566 | 0.166 | 0.088 | 0.118 | -0.018 | 0.094 |
|---|
| FANCA |   | 9 | 196 | 1 | 44 | 0.120 | -0.188 | 0.106 | -0.127 | -0.053 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| MNAT1 |   | 2 | 743 | 570 | 593 | 0.194 | 0.055 | 0.224 | 0.033 | 0.236 |
|---|
| ZHX1 |   | 5 | 360 | 1364 | 1328 | 0.094 | -0.187 | undef | undef | undef |
|---|
| FGFR2 |   | 7 | 256 | 179 | 201 | -0.024 | -0.001 | 0.129 | undef | 0.009 |
|---|
| HIF1A |   | 13 | 124 | 236 | 230 | -0.098 | 0.224 | -0.118 | 0.135 | 0.356 |
|---|
| IL6 |   | 9 | 196 | 366 | 365 | 0.257 | -0.106 | 0.226 | 0.024 | 0.035 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| PML |   | 6 | 301 | 478 | 473 | -0.068 | -0.010 | -0.007 | -0.153 | -0.070 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| DHFR |   | 4 | 440 | 179 | 209 | 0.019 | 0.029 | 0.232 | 0.129 | -0.050 |
|---|
| EPO |   | 4 | 440 | 1492 | 1461 | 0.109 | 0.086 | -0.083 | -0.048 | 0.155 |
|---|
| SNCAIP |   | 10 | 167 | 366 | 362 | -0.091 | 0.013 | 0.092 | 0.136 | 0.059 |
|---|
| HIPK3 |   | 9 | 196 | 1461 | 1417 | 0.069 | 0.005 | -0.027 | undef | 0.175 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| CCR5 |   | 2 | 743 | 412 | 439 | 0.153 | -0.276 | -0.159 | -0.156 | -0.067 |
|---|
| FOXA1 |   | 27 | 50 | 179 | 176 | -0.072 | 0.071 | -0.015 | 0.214 | 0.005 |
|---|
GO Enrichment output for subnetwork 6466 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.51E-09 | 3.69E-06 |
|---|
| glycine biosynthetic process | GO:0006545 |  | 1.391E-08 | 1.699E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2E-07 | 1.629E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.564E-07 | 4.009E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 7.576E-07 | 3.702E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 7.645E-07 | 3.113E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 8.366E-07 | 2.92E-04 |
|---|
| positive regulation of endothelial cell proliferation | GO:0001938 |  | 8.867E-07 | 2.708E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 1.225E-06 | 3.325E-04 |
|---|
| serine family amino acid biosynthetic process | GO:0009070 |  | 2.644E-06 | 6.458E-04 |
|---|
| negative regulation of growth | GO:0045926 |  | 3.138E-06 | 6.969E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.549E-09 | 6.132E-06 |
|---|
| glycine biosynthetic process | GO:0006545 |  | 1.827E-08 | 2.198E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.144E-07 | 2.522E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 6.689E-07 | 4.023E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 8.538E-07 | 4.108E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 9.678E-07 | 3.881E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.072E-06 | 3.683E-04 |
|---|
| interphase | GO:0051325 |  | 1.083E-06 | 3.258E-04 |
|---|
| positive regulation of endothelial cell proliferation | GO:0001938 |  | 1.162E-06 | 3.106E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 1.586E-06 | 3.815E-04 |
|---|
| serine family amino acid biosynthetic process | GO:0009070 |  | 2.486E-06 | 5.438E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.703E-09 | 1.542E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.417E-07 | 6.23E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 8.051E-07 | 6.173E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.179E-06 | 6.777E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.33E-06 | 6.116E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.728E-06 | 6.623E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 1.883E-06 | 6.188E-04 |
|---|
| interphase | GO:0051325 |  | 2.059E-06 | 5.92E-04 |
|---|
| positive regulation of endothelial cell proliferation | GO:0001938 |  | 2.576E-06 | 6.584E-04 |
|---|
| serine family amino acid biosynthetic process | GO:0009070 |  | 2.576E-06 | 5.926E-04 |
|---|
| glycine metabolic process | GO:0006544 |  | 7.641E-06 | 1.598E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.519E-07 | 2.799E-04 |
|---|
| positive regulation of endothelial cell proliferation | GO:0001938 |  | 1.06E-06 | 9.766E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.149E-06 | 7.061E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.463E-06 | 6.74E-04 |
|---|
| cytokine-mediated signaling pathway | GO:0019221 |  | 1.881E-06 | 6.933E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.881E-06 | 5.778E-04 |
|---|
| gland development | GO:0048732 |  | 7.461E-06 | 1.964E-03 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 8.031E-06 | 1.85E-03 |
|---|
| positive regulation of protein transport | GO:0051222 |  | 8.031E-06 | 1.645E-03 |
|---|
| hemoglobin biosynthetic process | GO:0042541 |  | 9.107E-06 | 1.678E-03 |
|---|
| prostate gland development | GO:0030850 |  | 9.107E-06 | 1.526E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.703E-09 | 1.542E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.417E-07 | 6.23E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 8.051E-07 | 6.173E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.179E-06 | 6.777E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.33E-06 | 6.116E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.728E-06 | 6.623E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 1.883E-06 | 6.188E-04 |
|---|
| interphase | GO:0051325 |  | 2.059E-06 | 5.92E-04 |
|---|
| positive regulation of endothelial cell proliferation | GO:0001938 |  | 2.576E-06 | 6.584E-04 |
|---|
| serine family amino acid biosynthetic process | GO:0009070 |  | 2.576E-06 | 5.926E-04 |
|---|
| glycine metabolic process | GO:0006544 |  | 7.641E-06 | 1.598E-03 |
|---|



