Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6447
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7557 | 3.400e-03 | 5.480e-04 | 7.658e-01 | 1.427e-06 |
|---|
| Loi | 0.2297 | 8.114e-02 | 1.159e-02 | 4.303e-01 | 4.048e-04 |
|---|
| Schmidt | 0.6726 | 0.000e+00 | 0.000e+00 | 4.581e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7906 | 0.000e+00 | 0.000e+00 | 3.488e-03 | 0.000e+00 |
|---|
| Wang | 0.2609 | 2.768e-03 | 4.463e-02 | 4.001e-01 | 4.943e-05 |
|---|
Expression data for subnetwork 6447 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
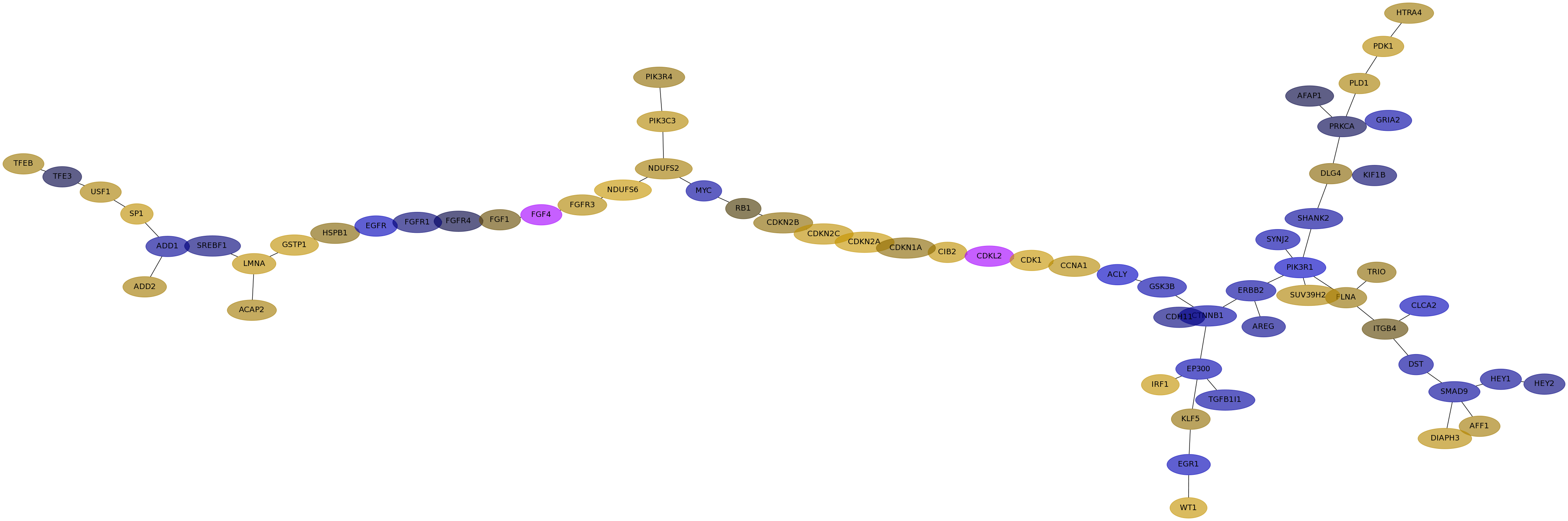
Score for each gene in subnetwork 6447 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| ADD1 |   | 8 | 222 | 366 | 373 | -0.085 | 0.116 | 0.035 | undef | -0.098 |
|---|
| CIB2 |   | 18 | 86 | 590 | 564 | 0.228 | -0.149 | 0.106 | 0.127 | 0.190 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| ADD2 |   | 8 | 222 | 614 | 599 | 0.113 | -0.022 | -0.061 | 0.270 | 0.180 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| DIAPH3 |   | 1 | 1195 | 2015 | 2022 | 0.179 | -0.125 | 0.000 | undef | undef |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| HEY2 |   | 8 | 222 | 570 | 555 | -0.045 | -0.021 | 0.398 | -0.024 | -0.058 |
|---|
| CDKN2C |   | 16 | 104 | 236 | 227 | 0.216 | -0.106 | 0.202 | -0.024 | -0.081 |
|---|
| DST |   | 22 | 69 | 1 | 25 | -0.094 | 0.343 | 0.065 | -0.062 | 0.194 |
|---|
| USF1 |   | 2 | 743 | 478 | 490 | 0.135 | 0.268 | undef | -0.019 | undef |
|---|
| ACAP2 |   | 1 | 1195 | 2015 | 2022 | 0.114 | 0.113 | 0.073 | 0.020 | 0.102 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| NDUFS2 |   | 1 | 1195 | 2015 | 2022 | 0.115 | 0.060 | -0.231 | 0.256 | 0.038 |
|---|
| PDK1 |   | 10 | 167 | 1 | 40 | 0.183 | -0.138 | -0.041 | -0.265 | -0.126 |
|---|
| SMAD9 |   | 9 | 196 | 296 | 285 | -0.090 | -0.153 | 0.055 | 0.081 | -0.059 |
|---|
| FGF4 |   | 2 | 743 | 882 | 892 | undef | undef | undef | 0.116 | undef |
|---|
| SHANK2 |   | 2 | 743 | 1083 | 1084 | -0.090 | -0.015 | 0.060 | undef | -0.111 |
|---|
| DLG4 |   | 5 | 360 | 806 | 784 | 0.059 | -0.113 | 0.151 | -0.131 | 0.123 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| FGFR1 |   | 26 | 52 | 638 | 615 | -0.036 | 0.200 | 0.087 | undef | -0.015 |
|---|
| FGF1 |   | 4 | 440 | 1604 | 1572 | 0.027 | undef | undef | 0.090 | undef |
|---|
| AREG |   | 5 | 360 | 1141 | 1109 | -0.067 | 0.112 | 0.076 | -0.173 | -0.054 |
|---|
| CLCA2 |   | 9 | 196 | 412 | 411 | -0.176 | 0.243 | -0.153 | 0.191 | -0.000 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| HTRA4 |   | 8 | 222 | 83 | 101 | 0.102 | undef | undef | undef | undef |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| HSPB1 |   | 22 | 69 | 1 | 25 | 0.053 | 0.169 | 0.017 | 0.179 | 0.161 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| HEY1 |   | 2 | 743 | 1867 | 1849 | -0.078 | -0.021 | 0.052 | 0.142 | 0.189 |
|---|
| CDKN2B |   | 7 | 256 | 236 | 237 | 0.066 | 0.050 | -0.028 | 0.039 | 0.153 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| NDUFS6 |   | 1 | 1195 | 2015 | 2022 | 0.261 | 0.068 | -0.021 | 0.236 | -0.038 |
|---|
| TRIO |   | 3 | 557 | 366 | 389 | 0.067 | 0.058 | -0.062 | 0.196 | -0.134 |
|---|
| PIK3R4 |   | 1 | 1195 | 2015 | 2022 | 0.073 | -0.038 | -0.050 | undef | 0.346 |
|---|
| CDH11 |   | 2 | 743 | 1222 | 1215 | -0.047 | 0.067 | -0.105 | undef | 0.074 |
|---|
| GRIA2 |   | 2 | 743 | 1492 | 1474 | -0.115 | -0.043 | 0.000 | -0.066 | 0.121 |
|---|
| GSTP1 |   | 14 | 117 | 1 | 34 | 0.222 | -0.166 | 0.201 | 0.126 | -0.136 |
|---|
| LMNA |   | 3 | 557 | 1047 | 1038 | 0.208 | 0.223 | -0.017 | 0.247 | 0.069 |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| FGFR3 |   | 9 | 196 | 179 | 195 | 0.160 | 0.071 | 0.018 | -0.050 | -0.111 |
|---|
| SUV39H2 |   | 8 | 222 | 318 | 325 | 0.150 | -0.097 | -0.057 | undef | 0.028 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| SREBF1 |   | 9 | 196 | 478 | 460 | -0.042 | 0.232 | -0.195 | 0.140 | -0.074 |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| AFF1 |   | 2 | 743 | 1598 | 1586 | 0.110 | 0.059 | -0.063 | 0.187 | 0.147 |
|---|
| SYNJ2 |   | 3 | 557 | 236 | 265 | -0.132 | 0.008 | 0.015 | undef | -0.066 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| FGFR4 |   | 21 | 73 | 179 | 183 | -0.011 | 0.188 | -0.089 | -0.018 | -0.049 |
|---|
| PIK3C3 |   | 1 | 1195 | 2015 | 2022 | 0.168 | 0.052 | 0.111 | 0.056 | 0.114 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| CDKL2 |   | 3 | 557 | 1260 | 1237 | undef | -0.129 | 0.000 | 0.109 | undef |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| TFEB |   | 1 | 1195 | 2015 | 2022 | 0.102 | 0.214 | -0.065 | undef | 0.024 |
|---|
| TFE3 |   | 1 | 1195 | 2015 | 2022 | -0.012 | 0.010 | -0.120 | -0.001 | 0.069 |
|---|
| ACLY |   | 6 | 301 | 1260 | 1224 | -0.236 | 0.071 | 0.009 | 0.158 | 0.115 |
|---|
| PLD1 |   | 17 | 95 | 1 | 29 | 0.132 | 0.033 | -0.158 | -0.062 | 0.123 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| KIF1B |   | 1 | 1195 | 2015 | 2022 | -0.029 | 0.071 | -0.192 | 0.085 | 0.019 |
|---|
| AFAP1 |   | 2 | 743 | 1492 | 1474 | -0.010 | -0.051 | -0.050 | undef | 0.052 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| IRF1 |   | 3 | 557 | 1629 | 1610 | 0.241 | -0.216 | -0.131 | -0.037 | -0.170 |
|---|
GO Enrichment output for subnetwork 6447 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 4.952E-11 | 1.21E-07 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 9.323E-11 | 1.139E-07 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.181E-10 | 1.776E-07 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 5.878E-09 | 3.59E-06 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 1.871E-08 | 9.142E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.22E-07 | 9.038E-05 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 3.051E-07 | 1.065E-04 |
|---|
| cellular response to nutrient levels | GO:0031669 |  | 3.82E-07 | 1.167E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.171E-07 | 2.218E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.395E-06 | 3.409E-04 |
|---|
| cellular response to nutrient | GO:0031670 |  | 1.714E-06 | 3.808E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 6.571E-11 | 1.581E-07 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.237E-10 | 1.488E-07 |
|---|
| cell cycle arrest | GO:0007050 |  | 1.365E-07 | 1.095E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.863E-07 | 1.121E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 2.303E-07 | 1.108E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.821E-07 | 1.131E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 3.111E-07 | 1.069E-04 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 6.824E-07 | 2.052E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.036E-06 | 2.771E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.48E-06 | 3.562E-04 |
|---|
| interphase | GO:0051325 |  | 1.826E-06 | 3.995E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.372E-10 | 5.456E-07 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 4.594E-10 | 5.284E-07 |
|---|
| cell cycle arrest | GO:0007050 |  | 2.456E-07 | 1.883E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 4.017E-07 | 2.31E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.326E-07 | 3.37E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 7.987E-07 | 3.062E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.061E-06 | 6.773E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.526E-06 | 1.014E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 4.005E-06 | 1.023E-03 |
|---|
| interphase | GO:0051325 |  | 4.115E-06 | 9.465E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 4.44E-06 | 9.283E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 2.045E-08 | 3.769E-05 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 6.94E-07 | 6.396E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.39E-06 | 8.542E-04 |
|---|
| cellular response to nutrient | GO:0031670 |  | 2.419E-06 | 1.115E-03 |
|---|
| cellular response to nutrient levels | GO:0031669 |  | 2.505E-06 | 9.232E-04 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 4.172E-06 | 1.282E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.047E-06 | 2.119E-03 |
|---|
| axon cargo transport | GO:0008088 |  | 8.392E-06 | 1.933E-03 |
|---|
| induction of an organ | GO:0001759 |  | 8.392E-06 | 1.719E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 1.311E-05 | 2.416E-03 |
|---|
| developmental induction | GO:0031128 |  | 1.337E-05 | 2.24E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.372E-10 | 5.456E-07 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 4.594E-10 | 5.284E-07 |
|---|
| cell cycle arrest | GO:0007050 |  | 2.456E-07 | 1.883E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 4.017E-07 | 2.31E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.326E-07 | 3.37E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 7.987E-07 | 3.062E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.061E-06 | 6.773E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.526E-06 | 1.014E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 4.005E-06 | 1.023E-03 |
|---|
| interphase | GO:0051325 |  | 4.115E-06 | 9.465E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 4.44E-06 | 9.283E-04 |
|---|



