Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6435
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7558 | 3.393e-03 | 5.470e-04 | 7.655e-01 | 1.421e-06 |
|---|
| Loi | 0.2298 | 8.100e-02 | 1.155e-02 | 4.300e-01 | 4.024e-04 |
|---|
| Schmidt | 0.6726 | 0.000e+00 | 0.000e+00 | 4.581e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7906 | 0.000e+00 | 0.000e+00 | 3.495e-03 | 0.000e+00 |
|---|
| Wang | 0.2607 | 2.794e-03 | 4.486e-02 | 4.012e-01 | 5.029e-05 |
|---|
Expression data for subnetwork 6435 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
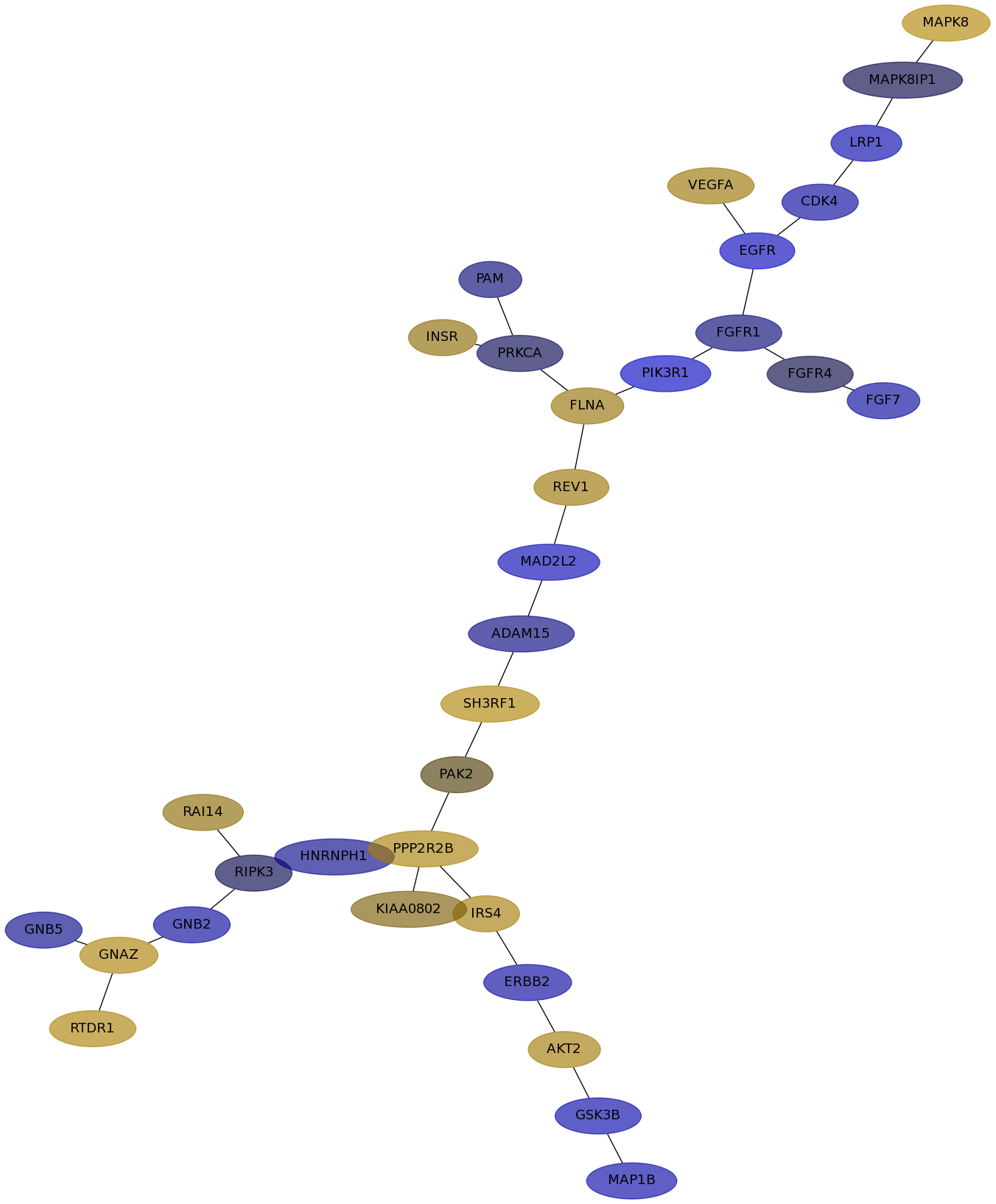
Score for each gene in subnetwork 6435 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| MAPK8 |   | 8 | 222 | 696 | 677 | 0.153 | 0.200 | 0.249 | -0.066 | 0.138 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| HNRNPH1 |   | 9 | 196 | 614 | 597 | -0.059 | 0.248 | 0.125 | 0.144 | -0.133 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| GNB2 |   | 11 | 148 | 525 | 505 | -0.091 | 0.082 | -0.062 | 0.208 | -0.062 |
|---|
| MAD2L2 |   | 2 | 743 | 1803 | 1782 | -0.167 | -0.009 | undef | 0.144 | undef |
|---|
| GNB5 |   | 9 | 196 | 1131 | 1100 | -0.059 | 0.026 | 0.064 | 0.284 | 0.103 |
|---|
| GNAZ |   | 15 | 111 | 478 | 457 | 0.134 | -0.116 | 0.308 | 0.267 | 0.011 |
|---|
| PAK2 |   | 26 | 52 | 179 | 177 | 0.014 | 0.161 | 0.081 | -0.044 | 0.062 |
|---|
| ADAM15 |   | 3 | 557 | 1803 | 1777 | -0.048 | -0.078 | -0.208 | 0.172 | -0.048 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| REV1 |   | 9 | 196 | 296 | 285 | 0.090 | -0.027 | 0.165 | 0.049 | 0.039 |
|---|
| FGFR4 |   | 21 | 73 | 179 | 183 | -0.011 | 0.188 | -0.089 | -0.018 | -0.049 |
|---|
| CDK4 |   | 4 | 440 | 913 | 904 | -0.092 | -0.201 | 0.091 | -0.043 | -0.017 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| RIPK3 |   | 19 | 85 | 179 | 186 | -0.014 | 0.040 | undef | 0.173 | undef |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| LRP1 |   | 2 | 743 | 1954 | 1933 | -0.138 | 0.026 | 0.119 | 0.063 | 0.112 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| FGFR1 |   | 26 | 52 | 638 | 615 | -0.036 | 0.200 | 0.087 | undef | -0.015 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAPK8IP1 |   | 4 | 440 | 1576 | 1543 | -0.012 | 0.058 | -0.020 | 0.282 | -0.049 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| SH3RF1 |   | 6 | 301 | 1222 | 1194 | 0.144 | 0.238 | undef | undef | undef |
|---|
| KIAA0802 |   | 3 | 557 | 2046 | 2009 | 0.041 | -0.123 | 0.123 | 0.059 | 0.037 |
|---|
| PAM |   | 10 | 167 | 638 | 621 | -0.035 | 0.307 | 0.200 | 0.002 | 0.104 |
|---|
| RAI14 |   | 2 | 743 | 1903 | 1878 | 0.063 | 0.188 | 0.114 | undef | 0.225 |
|---|
| FGF7 |   | 10 | 167 | 780 | 760 | -0.094 | 0.237 | 0.070 | 0.038 | 0.083 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| RTDR1 |   | 5 | 360 | 1364 | 1328 | 0.135 | 0.098 | 0.222 | 0.125 | undef |
|---|
GO Enrichment output for subnetwork 6435 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 7.964E-09 | 1.946E-05 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.982E-08 | 2.42E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 3.108E-08 | 2.531E-05 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 4.493E-07 | 2.744E-04 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 5.427E-07 | 2.652E-04 |
|---|
| regulation of behavior | GO:0050795 |  | 7.718E-07 | 3.143E-04 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 7.963E-07 | 2.779E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.175E-06 | 3.588E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.271E-06 | 3.451E-04 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 2.713E-06 | 6.628E-04 |
|---|
| chondrocyte differentiation | GO:0002062 |  | 3.723E-06 | 8.268E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 7.019E-09 | 1.689E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 2.74E-08 | 3.296E-05 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 3.25E-07 | 2.606E-04 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 3.964E-07 | 2.384E-04 |
|---|
| regulation of behavior | GO:0050795 |  | 5.733E-07 | 2.759E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 6.811E-07 | 2.731E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 7.243E-07 | 2.49E-04 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 7.267E-07 | 2.185E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.16E-06 | 3.102E-04 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 2.476E-06 | 5.958E-04 |
|---|
| chondrocyte differentiation | GO:0002062 |  | 3.398E-06 | 7.433E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.06E-08 | 2.439E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 4.958E-08 | 5.702E-05 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 5.653E-07 | 4.334E-04 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 7.104E-07 | 4.085E-04 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 8.752E-07 | 4.026E-04 |
|---|
| regulation of behavior | GO:0050795 |  | 1.067E-06 | 4.089E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.541E-06 | 5.064E-04 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 1.545E-06 | 4.442E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.466E-06 | 6.301E-04 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 3.689E-06 | 8.485E-04 |
|---|
| chondrocyte differentiation | GO:0002062 |  | 7.209E-06 | 1.507E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.491E-08 | 2.748E-05 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 3.547E-06 | 3.269E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 3.982E-06 | 2.446E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 3.982E-06 | 1.835E-03 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 4.924E-06 | 1.815E-03 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 5.299E-06 | 1.628E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 6.875E-06 | 1.81E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 8.733E-06 | 2.012E-03 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.244E-05 | 2.546E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.147E-05 | 5.8E-03 |
|---|
| regulation of MAP kinase activity | GO:0043405 |  | 4.09E-05 | 6.852E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.06E-08 | 2.439E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 4.958E-08 | 5.702E-05 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 5.653E-07 | 4.334E-04 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 7.104E-07 | 4.085E-04 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 8.752E-07 | 4.026E-04 |
|---|
| regulation of behavior | GO:0050795 |  | 1.067E-06 | 4.089E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.541E-06 | 5.064E-04 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 1.545E-06 | 4.442E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.466E-06 | 6.301E-04 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 3.689E-06 | 8.485E-04 |
|---|
| chondrocyte differentiation | GO:0002062 |  | 7.209E-06 | 1.507E-03 |
|---|



