Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6427
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7558 | 3.396e-03 | 5.470e-04 | 7.656e-01 | 1.422e-06 |
|---|
| Loi | 0.2298 | 8.106e-02 | 1.157e-02 | 4.301e-01 | 4.033e-04 |
|---|
| Schmidt | 0.6726 | 0.000e+00 | 0.000e+00 | 4.582e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7905 | 0.000e+00 | 0.000e+00 | 3.497e-03 | 0.000e+00 |
|---|
| Wang | 0.2608 | 2.788e-03 | 4.481e-02 | 4.010e-01 | 5.010e-05 |
|---|
Expression data for subnetwork 6427 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
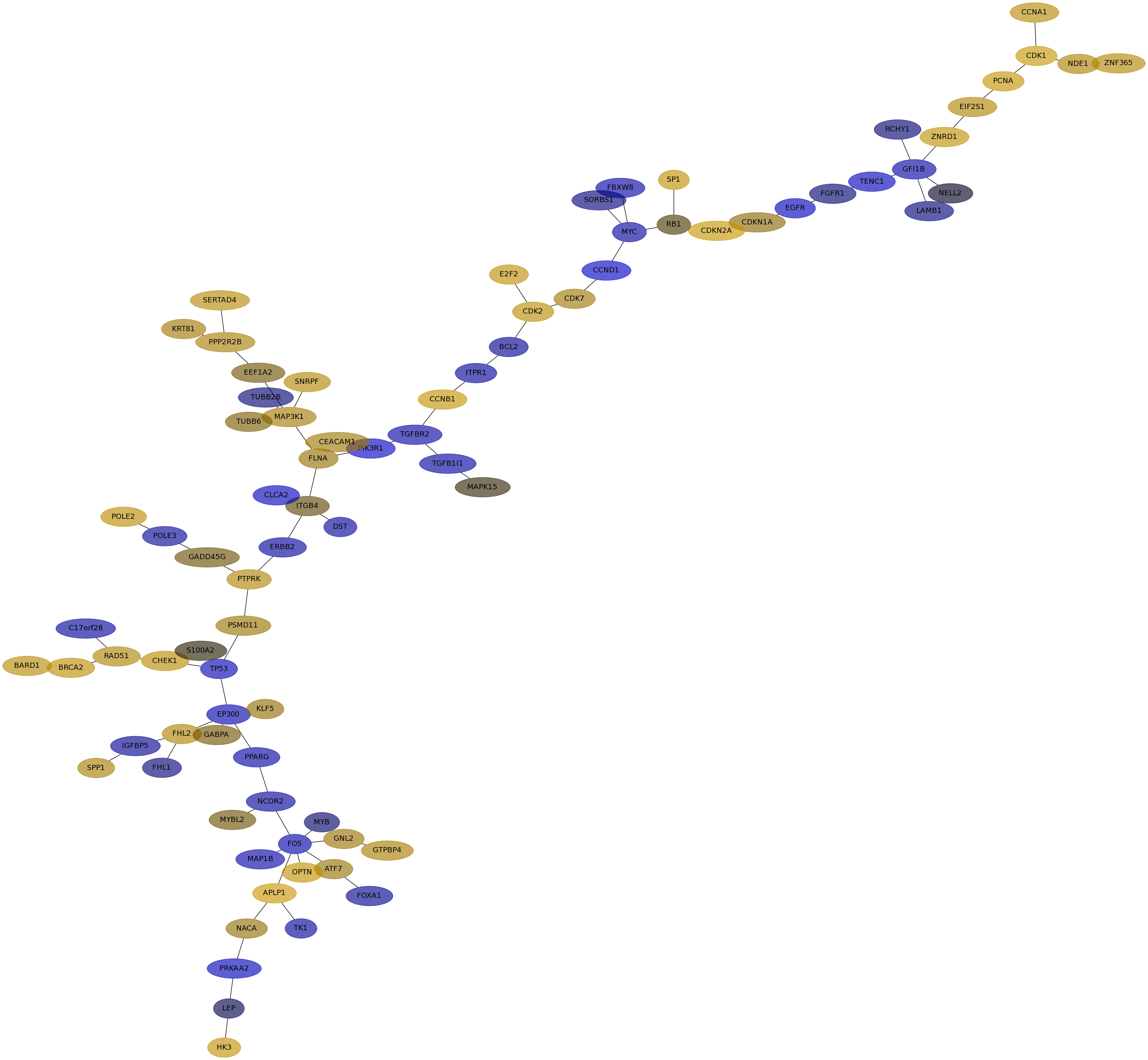
Score for each gene in subnetwork 6427 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| SNRPF |   | 2 | 743 | 478 | 490 | 0.163 | -0.066 | 0.064 | -0.116 | -0.103 |
|---|
| RCHY1 |   | 2 | 743 | 2080 | 2055 | -0.035 | 0.007 | 0.245 | undef | -0.026 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| TUBB6 |   | 3 | 557 | 1461 | 1440 | 0.045 | 0.114 | 0.187 | undef | 0.165 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| CCNB1 |   | 4 | 440 | 1116 | 1101 | 0.241 | -0.056 | 0.001 | undef | -0.043 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| DST |   | 22 | 69 | 1 | 25 | -0.094 | 0.343 | 0.065 | -0.062 | 0.194 |
|---|
| CCND1 |   | 26 | 52 | 1 | 24 | -0.258 | 0.074 | 0.117 | undef | 0.003 |
|---|
| NELL2 |   | 7 | 256 | 727 | 714 | -0.005 | 0.138 | 0.421 | -0.027 | 0.074 |
|---|
| PCNA |   | 15 | 111 | 1 | 32 | 0.243 | -0.158 | 0.027 | -0.099 | 0.003 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| POLE3 |   | 17 | 95 | 141 | 138 | -0.083 | -0.033 | 0.231 | 0.096 | 0.011 |
|---|
| ZNRD1 |   | 2 | 743 | 1364 | 1363 | 0.225 | -0.093 | undef | undef | undef |
|---|
| ZNF365 |   | 4 | 440 | 141 | 165 | 0.161 | 0.190 | -0.050 | -0.053 | -0.008 |
|---|
| CEACAM1 |   | 1 | 1195 | 2080 | 2087 | 0.102 | -0.039 | -0.106 | 0.049 | -0.014 |
|---|
| PTPRK |   | 14 | 117 | 83 | 96 | 0.156 | 0.141 | -0.094 | 0.048 | -0.024 |
|---|
| FBXW8 |   | 2 | 743 | 83 | 123 | -0.113 | 0.192 | undef | 0.219 | undef |
|---|
| GABPA |   | 23 | 66 | 696 | 665 | 0.033 | -0.183 | 0.066 | 0.141 | -0.161 |
|---|
| PPARG |   | 7 | 256 | 179 | 201 | -0.119 | 0.011 | 0.105 | 0.059 | 0.039 |
|---|
| SERTAD4 |   | 8 | 222 | 1268 | 1225 | 0.173 | 0.169 | undef | -0.050 | undef |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| CDK7 |   | 7 | 256 | 318 | 328 | 0.105 | 0.184 | 0.017 | 0.078 | -0.097 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| FGFR1 |   | 26 | 52 | 638 | 615 | -0.036 | 0.200 | 0.087 | undef | -0.015 |
|---|
| OPTN |   | 7 | 256 | 1 | 47 | 0.226 | -0.099 | 0.015 | undef | 0.035 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| CLCA2 |   | 9 | 196 | 412 | 411 | -0.176 | 0.243 | -0.153 | 0.191 | -0.000 |
|---|
| LAMB1 |   | 3 | 557 | 1077 | 1068 | -0.041 | 0.226 | -0.020 | -0.011 | 0.238 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| GFI1B |   | 1 | 1195 | 2080 | 2087 | -0.113 | 0.015 | 0.000 | -0.103 | undef |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| E2F2 |   | 13 | 124 | 1 | 35 | 0.212 | -0.047 | 0.162 | -0.131 | -0.092 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| PRKAA2 |   | 6 | 301 | 1165 | 1138 | -0.204 | 0.148 | 0.086 | 0.250 | -0.078 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| C17orf28 |   | 1 | 1195 | 2080 | 2087 | -0.094 | 0.094 | undef | undef | undef |
|---|
| NACA |   | 5 | 360 | 1165 | 1143 | 0.075 | 0.158 | 0.046 | -0.135 | 0.252 |
|---|
| ATF7 |   | 28 | 47 | 179 | 175 | 0.085 | 0.115 | -0.088 | 0.172 | -0.092 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| CHEK1 |   | 18 | 86 | 141 | 136 | 0.189 | -0.134 | 0.119 | -0.088 | -0.054 |
|---|
| GNL2 |   | 2 | 743 | 882 | 892 | 0.092 | 0.129 | -0.037 | -0.220 | -0.019 |
|---|
| KRT81 |   | 2 | 743 | 2080 | 2055 | 0.108 | -0.105 | 0.252 | 0.053 | 0.086 |
|---|
| TGFBR2 |   | 7 | 256 | 525 | 515 | -0.115 | -0.028 | 0.136 | 0.105 | -0.190 |
|---|
| S100A2 |   | 6 | 301 | 1 | 52 | 0.005 | 0.047 | 0.137 | -0.029 | -0.008 |
|---|
| SPP1 |   | 26 | 52 | 83 | 91 | 0.130 | 0.119 | 0.184 | 0.204 | 0.153 |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| APLP1 |   | 8 | 222 | 727 | 703 | 0.284 | 0.043 | -0.105 | -0.121 | -0.190 |
|---|
| MAPK15 |   | 1 | 1195 | 2080 | 2087 | 0.007 | -0.048 | undef | undef | undef |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| EEF1A2 |   | 8 | 222 | 457 | 450 | 0.032 | 0.220 | -0.119 | 0.018 | 0.102 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| LEP |   | 2 | 743 | 1364 | 1363 | -0.014 | -0.019 | 0.145 | -0.079 | 0.059 |
|---|
| PSMD11 |   | 6 | 301 | 1437 | 1408 | 0.088 | -0.090 | -0.137 | 0.060 | -0.078 |
|---|
| HK3 |   | 2 | 743 | 1364 | 1363 | 0.226 | -0.230 | -0.046 | -0.132 | -0.081 |
|---|
| MYB |   | 28 | 47 | 366 | 344 | -0.028 | 0.075 | 0.002 | 0.065 | -0.012 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| MAP3K1 |   | 5 | 360 | 141 | 155 | 0.112 | 0.039 | undef | undef | undef |
|---|
| TK1 |   | 4 | 440 | 727 | 723 | -0.089 | -0.156 | 0.029 | 0.119 | -0.052 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| FHL2 |   | 21 | 73 | 658 | 630 | 0.143 | 0.239 | -0.033 | 0.107 | 0.294 |
|---|
| GTPBP4 |   | 4 | 440 | 882 | 870 | 0.129 | -0.179 | 0.076 | -0.043 | -0.026 |
|---|
| MYBL2 |   | 4 | 440 | 1604 | 1572 | 0.031 | -0.220 | 0.034 | -0.034 | 0.006 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| TENC1 |   | 2 | 743 | 1492 | 1474 | -0.194 | 0.149 | 0.093 | undef | -0.159 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| POLE2 |   | 4 | 440 | 727 | 723 | 0.200 | -0.203 | 0.049 | 0.000 | -0.010 |
|---|
| RAD51 |   | 13 | 124 | 727 | 699 | 0.146 | -0.098 | 0.135 | -0.070 | 0.061 |
|---|
| ITPR1 |   | 4 | 440 | 1576 | 1543 | -0.096 | 0.077 | 0.015 | undef | 0.008 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| EIF2S1 |   | 1 | 1195 | 2080 | 2087 | 0.153 | -0.055 | 0.021 | -0.011 | 0.110 |
|---|
| TUBB2B |   | 13 | 124 | 141 | 141 | -0.036 | 0.304 | 0.045 | 0.187 | 0.139 |
|---|
| FHL1 |   | 14 | 117 | 614 | 591 | -0.041 | 0.104 | 0.301 | undef | 0.266 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| NDE1 |   | 3 | 557 | 141 | 170 | 0.142 | -0.058 | 0.142 | 0.107 | 0.111 |
|---|
| CDK2 |   | 31 | 43 | 179 | 174 | 0.196 | -0.096 | 0.109 | 0.001 | 0.362 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| FOXA1 |   | 27 | 50 | 179 | 176 | -0.072 | 0.071 | -0.015 | 0.214 | 0.005 |
|---|
GO Enrichment output for subnetwork 6427 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.541E-14 | 6.207E-11 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.197E-11 | 6.349E-08 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 1.432E-09 | 1.166E-06 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 6.385E-09 | 3.9E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.001E-08 | 4.89E-06 |
|---|
| response to UV | GO:0009411 |  | 3.627E-08 | 1.477E-05 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.196E-07 | 4.175E-05 |
|---|
| G1 phase | GO:0051318 |  | 1.665E-07 | 5.086E-05 |
|---|
| meiosis | GO:0007126 |  | 5.458E-07 | 1.482E-04 |
|---|
| meiotic cell cycle | GO:0051321 |  | 5.938E-07 | 1.451E-04 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.005E-06 | 2.232E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.781E-12 | 4.286E-09 |
|---|
| interphase | GO:0051325 |  | 2.602E-12 | 3.13E-09 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.432E-10 | 1.149E-07 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 2.854E-09 | 1.716E-06 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.268E-08 | 6.103E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.204E-08 | 8.836E-06 |
|---|
| response to UV | GO:0009411 |  | 6.102E-08 | 2.097E-05 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 2.866E-07 | 8.618E-05 |
|---|
| cell cycle arrest | GO:0007050 |  | 3.628E-07 | 9.698E-05 |
|---|
| meiosis | GO:0007126 |  | 6.333E-07 | 1.524E-04 |
|---|
| meiotic cell cycle | GO:0051321 |  | 6.945E-07 | 1.519E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.563E-10 | 3.595E-07 |
|---|
| interphase | GO:0051325 |  | 2.015E-10 | 2.318E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.021E-08 | 7.829E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.748E-08 | 3.88E-05 |
|---|
| response to UV | GO:0009411 |  | 1.097E-07 | 5.048E-05 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 3.186E-07 | 1.221E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 6.056E-07 | 1.99E-04 |
|---|
| cell cycle arrest | GO:0007050 |  | 6.63E-07 | 1.906E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 7.413E-07 | 1.895E-04 |
|---|
| response to gamma radiation | GO:0010332 |  | 2.316E-06 | 5.327E-04 |
|---|
| regulation of DNA replication | GO:0006275 |  | 3.044E-06 | 6.366E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.601E-07 | 2.951E-04 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.23E-06 | 1.134E-03 |
|---|
| G1 phase | GO:0051318 |  | 1.995E-06 | 1.226E-03 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 2.963E-06 | 1.365E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 3.985E-06 | 1.469E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 4.706E-06 | 1.446E-03 |
|---|
| T cell lineage commitment | GO:0002360 |  | 5.33E-06 | 1.403E-03 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 7.562E-06 | 1.742E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.767E-06 | 1.795E-03 |
|---|
| focal adhesion formation | GO:0048041 |  | 1.06E-05 | 1.953E-03 |
|---|
| meiosis | GO:0007126 |  | 1.162E-05 | 1.948E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.563E-10 | 3.595E-07 |
|---|
| interphase | GO:0051325 |  | 2.015E-10 | 2.318E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.021E-08 | 7.829E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.748E-08 | 3.88E-05 |
|---|
| response to UV | GO:0009411 |  | 1.097E-07 | 5.048E-05 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 3.186E-07 | 1.221E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 6.056E-07 | 1.99E-04 |
|---|
| cell cycle arrest | GO:0007050 |  | 6.63E-07 | 1.906E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 7.413E-07 | 1.895E-04 |
|---|
| response to gamma radiation | GO:0010332 |  | 2.316E-06 | 5.327E-04 |
|---|
| regulation of DNA replication | GO:0006275 |  | 3.044E-06 | 6.366E-04 |
|---|



