Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6422
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7559 | 3.391e-03 | 5.460e-04 | 7.655e-01 | 1.417e-06 |
|---|
| Loi | 0.2297 | 8.128e-02 | 1.163e-02 | 4.307e-01 | 4.071e-04 |
|---|
| Schmidt | 0.6727 | 0.000e+00 | 0.000e+00 | 4.573e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7905 | 0.000e+00 | 0.000e+00 | 3.499e-03 | 0.000e+00 |
|---|
| Wang | 0.2608 | 2.791e-03 | 4.483e-02 | 4.011e-01 | 5.018e-05 |
|---|
Expression data for subnetwork 6422 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
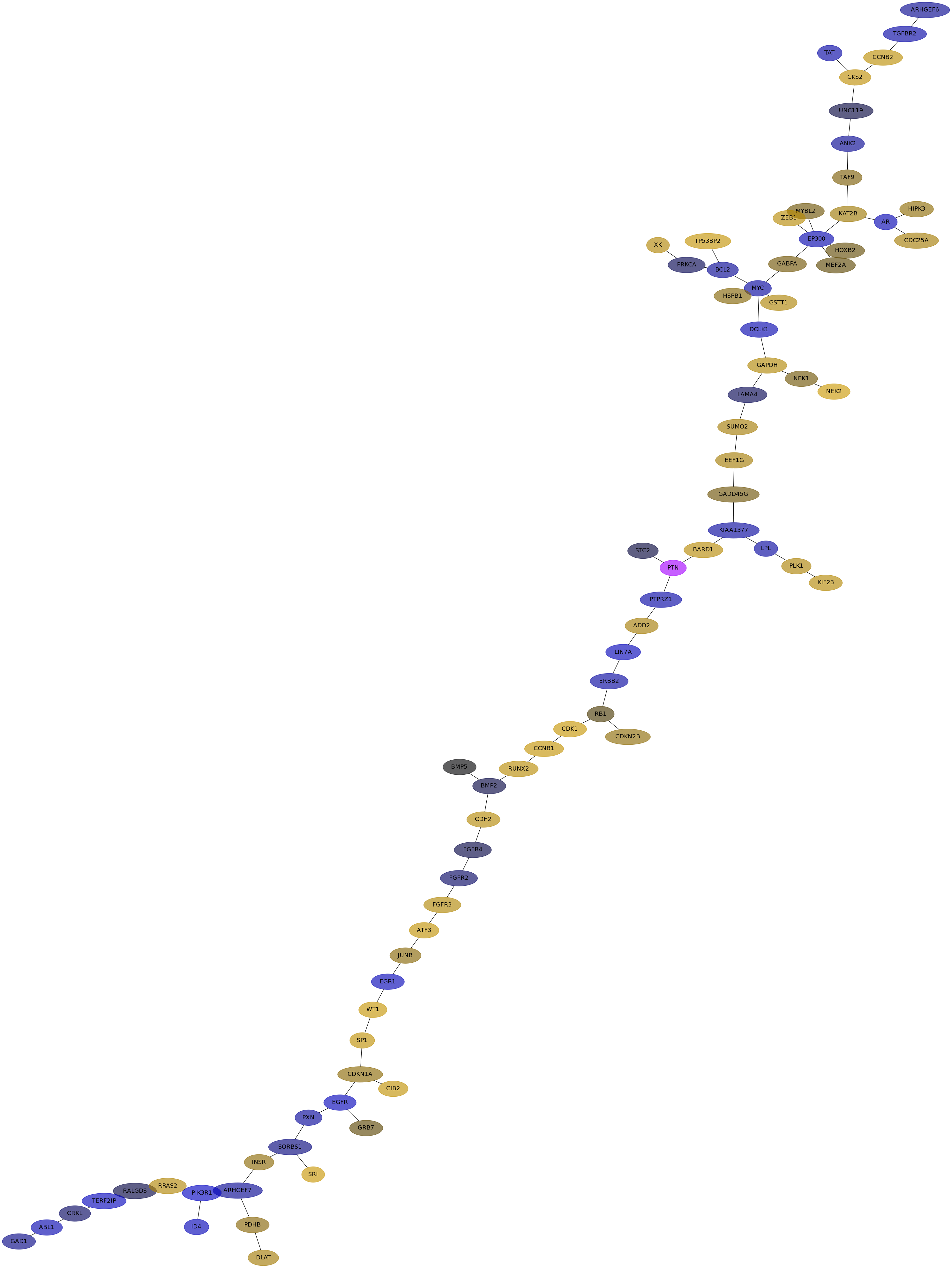
Score for each gene in subnetwork 6422 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| SUMO2 |   | 22 | 69 | 366 | 345 | 0.117 | 0.062 | 0.294 | 0.141 | -0.096 |
|---|
| CIB2 |   | 18 | 86 | 590 | 564 | 0.228 | -0.149 | 0.106 | 0.127 | 0.190 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| ADD2 |   | 8 | 222 | 614 | 599 | 0.113 | -0.022 | -0.061 | 0.270 | 0.180 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| CCNB1 |   | 4 | 440 | 1116 | 1101 | 0.241 | -0.056 | 0.001 | undef | -0.043 |
|---|
| EEF1G |   | 8 | 222 | 882 | 869 | 0.116 | -0.224 | 0.131 | 0.086 | 0.082 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| TERF2IP |   | 1 | 1195 | 2100 | 2105 | -0.201 | 0.128 | -0.042 | 0.124 | 0.087 |
|---|
| CRKL |   | 6 | 301 | 780 | 764 | -0.023 | 0.056 | 0.271 | -0.025 | -0.008 |
|---|
| SRI |   | 7 | 256 | 614 | 601 | 0.270 | -0.040 | 0.181 | 0.185 | 0.057 |
|---|
| ZEB1 |   | 2 | 743 | 1564 | 1549 | 0.178 | -0.158 | -0.094 | undef | -0.158 |
|---|
| LIN7A |   | 6 | 301 | 366 | 377 | -0.189 | -0.074 | 0.108 | 0.065 | -0.146 |
|---|
| TP53BP2 |   | 11 | 148 | 692 | 668 | 0.245 | -0.012 | 0.129 | 0.021 | 0.017 |
|---|
| CKS2 |   | 9 | 196 | 928 | 903 | 0.214 | -0.220 | 0.089 | -0.003 | -0.056 |
|---|
| PLK1 |   | 11 | 148 | 236 | 232 | 0.142 | -0.143 | 0.094 | -0.192 | -0.163 |
|---|
| TAT |   | 11 | 148 | 83 | 97 | -0.113 | 0.030 | -0.182 | 0.037 | -0.074 |
|---|
| LAMA4 |   | 13 | 124 | 696 | 670 | -0.016 | 0.138 | 0.209 | 0.172 | 0.343 |
|---|
| GAPDH |   | 11 | 148 | 141 | 143 | 0.159 | -0.040 | -0.053 | 0.088 | 0.029 |
|---|
| GABPA |   | 23 | 66 | 696 | 665 | 0.033 | -0.183 | 0.066 | 0.141 | -0.161 |
|---|
| ANK2 |   | 3 | 557 | 1604 | 1577 | -0.073 | -0.023 | 0.095 | -0.019 | 0.095 |
|---|
| ATF3 |   | 17 | 95 | 179 | 189 | 0.225 | -0.021 | 0.000 | undef | -0.039 |
|---|
| PDHB |   | 13 | 124 | 525 | 503 | 0.056 | 0.017 | 0.197 | -0.205 | 0.193 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| ARHGEF6 |   | 10 | 167 | 525 | 507 | -0.068 | -0.120 | 0.166 | undef | -0.115 |
|---|
| DCLK1 |   | 7 | 256 | 141 | 151 | -0.145 | -0.006 | -0.048 | 0.188 | -0.035 |
|---|
| LPL |   | 13 | 124 | 236 | 230 | -0.102 | -0.062 | 0.292 | 0.087 | -0.105 |
|---|
| TAF9 |   | 2 | 743 | 1165 | 1163 | 0.043 | -0.014 | undef | undef | undef |
|---|
| HSPB1 |   | 22 | 69 | 1 | 25 | 0.053 | 0.169 | 0.017 | 0.179 | 0.161 |
|---|
| PTN |   | 24 | 62 | 236 | 225 | undef | 0.022 | 0.000 | 0.054 | -0.092 |
|---|
| DLAT |   | 4 | 440 | 1141 | 1127 | 0.111 | 0.029 | 0.072 | undef | -0.014 |
|---|
| CDKN2B |   | 7 | 256 | 236 | 237 | 0.066 | 0.050 | -0.028 | 0.039 | 0.153 |
|---|
| RUNX2 |   | 2 | 743 | 1116 | 1121 | 0.184 | -0.104 | -0.127 | 0.013 | undef |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| KAT2B |   | 13 | 124 | 318 | 316 | 0.102 | -0.157 | -0.091 | 0.136 | -0.087 |
|---|
| ARHGEF7 |   | 25 | 59 | 83 | 92 | -0.074 | -0.061 | 0.208 | 0.028 | 0.015 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| GSTT1 |   | 15 | 111 | 1 | 32 | 0.148 | 0.107 | -0.190 | 0.022 | -0.064 |
|---|
| KIF23 |   | 4 | 440 | 957 | 942 | 0.168 | -0.063 | 0.162 | undef | 0.033 |
|---|
| RALGDS |   | 5 | 360 | 2024 | 1972 | -0.011 | 0.199 | 0.033 | undef | -0.009 |
|---|
| MEF2A |   | 4 | 440 | 614 | 618 | 0.022 | 0.036 | 0.265 | -0.094 | 0.124 |
|---|
| KIAA1377 |   | 18 | 86 | 141 | 136 | -0.095 | 0.116 | undef | 0.198 | undef |
|---|
| TGFBR2 |   | 7 | 256 | 525 | 515 | -0.115 | -0.028 | 0.136 | 0.105 | -0.190 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| ABL1 |   | 13 | 124 | 727 | 699 | -0.152 | 0.200 | 0.108 | undef | -0.059 |
|---|
| BMP2 |   | 11 | 148 | 525 | 505 | -0.011 | 0.097 | 0.148 | -0.114 | -0.069 |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| FGFR3 |   | 9 | 196 | 179 | 195 | 0.160 | 0.071 | 0.018 | -0.050 | -0.111 |
|---|
| UNC119 |   | 10 | 167 | 83 | 98 | -0.009 | -0.324 | -0.099 | 0.094 | 0.071 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| JUNB |   | 20 | 81 | 179 | 185 | 0.057 | 0.086 | 0.065 | 0.001 | -0.015 |
|---|
| STC2 |   | 5 | 360 | 236 | 242 | -0.009 | 0.056 | 0.181 | -0.185 | 0.016 |
|---|
| NEK2 |   | 8 | 222 | 141 | 149 | 0.292 | -0.106 | 0.020 | 0.083 | 0.165 |
|---|
| PTPRZ1 |   | 9 | 196 | 570 | 554 | -0.114 | 0.183 | 0.233 | -0.316 | -0.095 |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| GAD1 |   | 1 | 1195 | 2100 | 2105 | -0.055 | 0.151 | 0.114 | -0.094 | -0.029 |
|---|
| RRAS2 |   | 7 | 256 | 1681 | 1633 | 0.155 | 0.076 | 0.289 | 0.044 | 0.031 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| FGFR4 |   | 21 | 73 | 179 | 183 | -0.011 | 0.188 | -0.089 | -0.018 | -0.049 |
|---|
| CDC25A |   | 8 | 222 | 570 | 555 | 0.113 | -0.170 | 0.151 | -0.301 | -0.003 |
|---|
| PXN |   | 12 | 140 | 1 | 36 | -0.088 | 0.284 | -0.022 | 0.060 | 0.114 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| MYBL2 |   | 4 | 440 | 1604 | 1572 | 0.031 | -0.220 | 0.034 | -0.034 | 0.006 |
|---|
| XK |   | 18 | 86 | 179 | 187 | 0.154 | -0.000 | 0.067 | -0.123 | -0.040 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| BMP5 |   | 1 | 1195 | 2100 | 2105 | -0.000 | -0.011 | 0.011 | 0.047 | -0.082 |
|---|
| NEK1 |   | 15 | 111 | 141 | 139 | 0.032 | 0.184 | 0.049 | undef | 0.135 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| FGFR2 |   | 7 | 256 | 179 | 201 | -0.024 | -0.001 | 0.129 | undef | 0.009 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| CCNB2 |   | 4 | 440 | 842 | 831 | 0.206 | -0.119 | 0.004 | -0.006 | 0.134 |
|---|
| CDH2 |   | 10 | 167 | 727 | 702 | 0.169 | 0.109 | -0.016 | 0.123 | 0.155 |
|---|
| HIPK3 |   | 9 | 196 | 1461 | 1417 | 0.069 | 0.005 | -0.027 | undef | 0.175 |
|---|
GO Enrichment output for subnetwork 6422 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 8.346E-09 | 2.039E-05 |
|---|
| cartilage development | GO:0051216 |  | 3.067E-08 | 3.747E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.98E-07 | 1.612E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.188E-07 | 1.337E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.856E-07 | 1.396E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 2.892E-07 | 1.178E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.892E-07 | 1.009E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 3.219E-07 | 9.831E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.31E-07 | 8.984E-05 |
|---|
| chondrocyte differentiation | GO:0002062 |  | 4.003E-07 | 9.78E-05 |
|---|
| thymus development | GO:0048538 |  | 4.003E-07 | 8.891E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.105E-08 | 2.658E-05 |
|---|
| interphase | GO:0051325 |  | 1.455E-08 | 1.751E-05 |
|---|
| cartilage development | GO:0051216 |  | 4.172E-08 | 3.346E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 2.501E-07 | 1.504E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.773E-07 | 1.334E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 3.207E-07 | 1.286E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 3.95E-07 | 1.358E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 4.123E-07 | 1.24E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.239E-07 | 1.133E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.849E-07 | 1.167E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 4.912E-07 | 1.074E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.644E-08 | 8.38E-05 |
|---|
| interphase | GO:0051325 |  | 4.464E-08 | 5.134E-05 |
|---|
| cartilage development | GO:0051216 |  | 1.43E-07 | 1.096E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 4.165E-07 | 2.395E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 4.704E-07 | 2.164E-04 |
|---|
| thymus development | GO:0048538 |  | 5.388E-07 | 2.065E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 6.687E-07 | 2.197E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 7.338E-07 | 2.11E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 8.574E-07 | 2.191E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.056E-06 | 2.429E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.325E-06 | 2.771E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| thymus development | GO:0048538 |  | 4.656E-08 | 8.58E-05 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 2.31E-06 | 2.128E-03 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 7.039E-06 | 4.324E-03 |
|---|
| focal adhesion formation | GO:0048041 |  | 8.495E-06 | 3.914E-03 |
|---|
| regulation of embryonic development | GO:0045995 |  | 8.495E-06 | 3.131E-03 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.056E-05 | 3.245E-03 |
|---|
| regulation of cell migration | GO:0030334 |  | 1.527E-05 | 4.02E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.705E-05 | 3.928E-03 |
|---|
| T cell homeostasis | GO:0043029 |  | 2.353E-05 | 4.818E-03 |
|---|
| regulation of locomotion | GO:0040012 |  | 2.827E-05 | 5.21E-03 |
|---|
| regulation of cell motion | GO:0051270 |  | 2.827E-05 | 4.736E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.644E-08 | 8.38E-05 |
|---|
| interphase | GO:0051325 |  | 4.464E-08 | 5.134E-05 |
|---|
| cartilage development | GO:0051216 |  | 1.43E-07 | 1.096E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 4.165E-07 | 2.395E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 4.704E-07 | 2.164E-04 |
|---|
| thymus development | GO:0048538 |  | 5.388E-07 | 2.065E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 6.687E-07 | 2.197E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 7.338E-07 | 2.11E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 8.574E-07 | 2.191E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.056E-06 | 2.429E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.325E-06 | 2.771E-04 |
|---|



