Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6410
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7561 | 3.377e-03 | 5.430e-04 | 7.648e-01 | 1.403e-06 |
|---|
| Loi | 0.2296 | 8.140e-02 | 1.166e-02 | 4.310e-01 | 4.091e-04 |
|---|
| Schmidt | 0.6726 | 0.000e+00 | 0.000e+00 | 4.577e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7905 | 0.000e+00 | 0.000e+00 | 3.501e-03 | 0.000e+00 |
|---|
| Wang | 0.2606 | 2.813e-03 | 4.504e-02 | 4.020e-01 | 5.093e-05 |
|---|
Expression data for subnetwork 6410 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
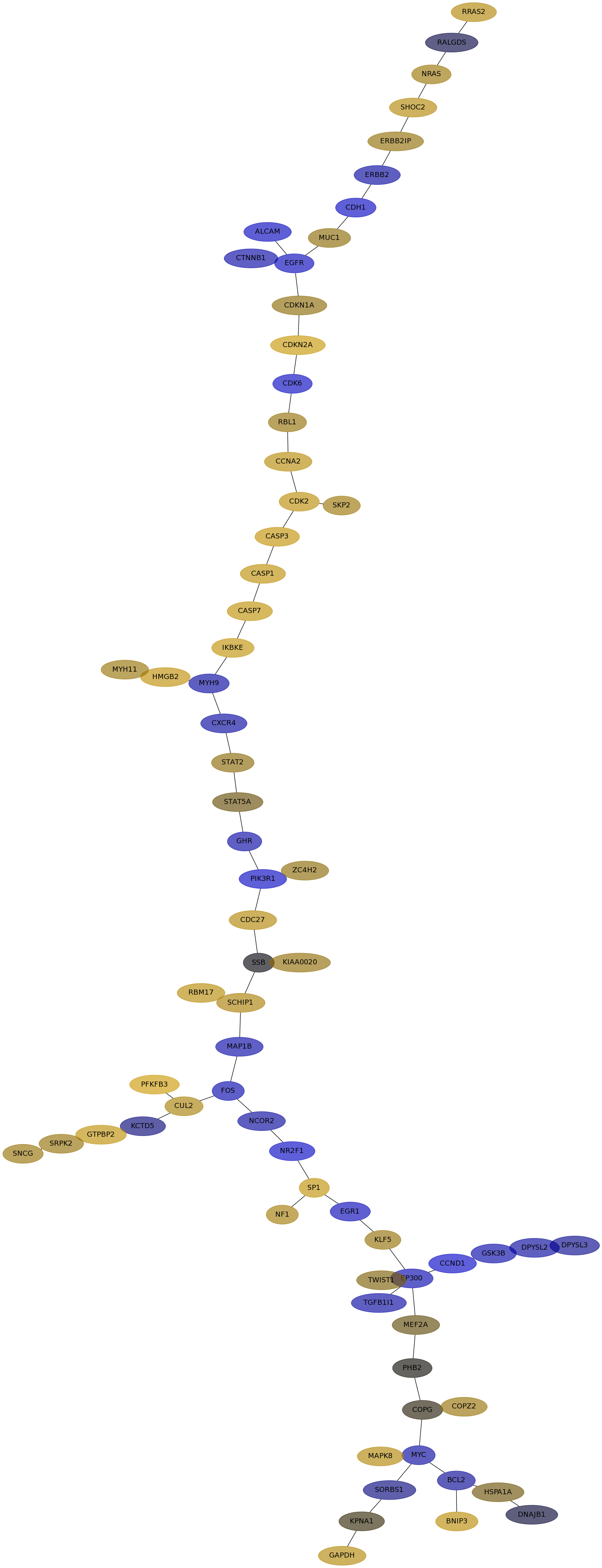
Score for each gene in subnetwork 6410 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| MAPK8 |   | 8 | 222 | 696 | 677 | 0.153 | 0.200 | 0.249 | -0.066 | 0.138 |
|---|
| CASP3 |   | 11 | 148 | 141 | 143 | 0.234 | -0.032 | -0.142 | -0.097 | -0.084 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| NF1 |   | 9 | 196 | 552 | 541 | 0.110 | -0.052 | -0.032 | 0.090 | -0.094 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| MYH9 |   | 4 | 440 | 1131 | 1108 | -0.103 | 0.227 | 0.007 | -0.150 | 0.165 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| KCTD5 |   | 1 | 1195 | 2163 | 2167 | -0.038 | -0.128 | -0.143 | -0.073 | 0.060 |
|---|
| HSPA1A |   | 2 | 743 | 648 | 666 | 0.030 | 0.164 | 0.070 | -0.023 | 0.046 |
|---|
| CCND1 |   | 26 | 52 | 1 | 24 | -0.258 | 0.074 | 0.117 | undef | 0.003 |
|---|
| CCNA2 |   | 21 | 73 | 179 | 183 | 0.176 | -0.053 | 0.188 | -0.078 | 0.246 |
|---|
| GTPBP2 |   | 2 | 743 | 1203 | 1206 | 0.211 | 0.033 | -0.132 | 0.119 | -0.002 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| DPYSL2 |   | 1 | 1195 | 2163 | 2167 | -0.095 | -0.037 | 0.296 | 0.051 | 0.104 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| CDK6 |   | 16 | 104 | 318 | 314 | -0.229 | 0.107 | -0.028 | -0.118 | 0.104 |
|---|
| SRPK2 |   | 3 | 557 | 1203 | 1187 | 0.076 | 0.183 | -0.087 | undef | 0.054 |
|---|
| SHOC2 |   | 1 | 1195 | 2163 | 2167 | 0.162 | -0.084 | 0.189 | 0.071 | 0.045 |
|---|
| GAPDH |   | 11 | 148 | 141 | 143 | 0.159 | -0.040 | -0.053 | 0.088 | 0.029 |
|---|
| NRAS |   | 2 | 743 | 2090 | 2082 | 0.090 | -0.211 | -0.054 | 0.076 | 0.021 |
|---|
| PFKFB3 |   | 3 | 557 | 1659 | 1632 | 0.301 | 0.085 | 0.328 | 0.056 | 0.331 |
|---|
| ERBB2IP |   | 1 | 1195 | 2163 | 2167 | 0.073 | 0.160 | 0.087 | -0.036 | 0.200 |
|---|
| ALCAM |   | 8 | 222 | 1 | 46 | -0.208 | 0.193 | 0.067 | 0.088 | -0.010 |
|---|
| KIAA0020 |   | 2 | 743 | 1141 | 1144 | 0.071 | -0.112 | 0.018 | -0.140 | -0.017 |
|---|
| STAT2 |   | 1 | 1195 | 2163 | 2167 | 0.061 | -0.008 | 0.000 | 0.176 | 0.155 |
|---|
| MYH11 |   | 2 | 743 | 2163 | 2145 | 0.084 | -0.003 | 0.211 | undef | 0.094 |
|---|
| DPYSL3 |   | 1 | 1195 | 2163 | 2167 | -0.065 | 0.133 | 0.021 | 0.125 | 0.261 |
|---|
| MUC1 |   | 13 | 124 | 1547 | 1490 | 0.063 | 0.214 | -0.190 | undef | -0.132 |
|---|
| RBL1 |   | 21 | 73 | 1 | 27 | 0.078 | undef | undef | 0.051 | undef |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| SSB |   | 2 | 743 | 1803 | 1782 | -0.001 | -0.058 | 0.147 | -0.071 | -0.161 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| COPG |   | 5 | 360 | 570 | 562 | 0.004 | 0.080 | -0.158 | undef | 0.280 |
|---|
| SNCG |   | 3 | 557 | 236 | 265 | 0.080 | -0.086 | 0.088 | -0.090 | -0.226 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| COPZ2 |   | 2 | 743 | 1629 | 1614 | 0.081 | 0.013 | 0.127 | 0.126 | 0.123 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| PHB2 |   | 2 | 743 | 1364 | 1363 | 0.001 | -0.154 | 0.071 | -0.107 | -0.054 |
|---|
| RALGDS |   | 5 | 360 | 2024 | 1972 | -0.011 | 0.199 | 0.033 | undef | -0.009 |
|---|
| MEF2A |   | 4 | 440 | 614 | 618 | 0.022 | 0.036 | 0.265 | -0.094 | 0.124 |
|---|
| DNAJB1 |   | 1 | 1195 | 2163 | 2167 | -0.007 | 0.052 | 0.039 | 0.054 | -0.007 |
|---|
| RBM17 |   | 1 | 1195 | 2163 | 2167 | 0.174 | -0.322 | undef | undef | undef |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| SCHIP1 |   | 10 | 167 | 478 | 459 | 0.130 | 0.136 | 0.105 | -0.022 | 0.198 |
|---|
| BNIP3 |   | 11 | 148 | 236 | 232 | 0.193 | 0.071 | 0.102 | 0.119 | 0.285 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| RRAS2 |   | 7 | 256 | 1681 | 1633 | 0.155 | 0.076 | 0.289 | 0.044 | 0.031 |
|---|
| CASP1 |   | 6 | 301 | 552 | 544 | 0.204 | -0.166 | -0.046 | undef | -0.034 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| ZC4H2 |   | 17 | 95 | 570 | 546 | 0.065 | 0.061 | 0.304 | -0.072 | 0.059 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| SKP2 |   | 29 | 45 | 1 | 23 | 0.092 | -0.043 | 0.237 | undef | -0.009 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| NR2F1 |   | 25 | 59 | 83 | 92 | -0.252 | 0.113 | 0.133 | -0.116 | 0.207 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| CASP7 |   | 6 | 301 | 614 | 602 | 0.216 | 0.095 | 0.255 | undef | 0.035 |
|---|
| CXCR4 |   | 2 | 743 | 882 | 892 | -0.103 | -0.189 | -0.094 | -0.160 | -0.010 |
|---|
| KPNA1 |   | 9 | 196 | 141 | 147 | 0.007 | 0.152 | -0.083 | 0.073 | 0.086 |
|---|
| IKBKE |   | 10 | 167 | 552 | 539 | 0.227 | -0.041 | -0.095 | -0.197 | 0.079 |
|---|
| CDC27 |   | 6 | 301 | 1010 | 986 | 0.156 | -0.086 | -0.029 | 0.006 | 0.348 |
|---|
| CUL2 |   | 7 | 256 | 842 | 816 | 0.124 | -0.192 | -0.090 | -0.035 | 0.166 |
|---|
| HMGB2 |   | 2 | 743 | 2163 | 2145 | 0.214 | -0.085 | 0.151 | undef | -0.015 |
|---|
| STAT5A |   | 1 | 1195 | 2163 | 2167 | 0.025 | -0.086 | -0.174 | -0.256 | -0.072 |
|---|
| CDK2 |   | 31 | 43 | 179 | 174 | 0.196 | -0.096 | 0.109 | 0.001 | 0.362 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
GO Enrichment output for subnetwork 6410 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.682E-10 | 4.109E-07 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.995E-10 | 2.436E-07 |
|---|
| response to UV | GO:0009411 |  | 7.521E-10 | 6.124E-07 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 2.914E-08 | 1.78E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 6.297E-08 | 3.076E-05 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 7.202E-08 | 2.933E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 9.411E-08 | 3.284E-05 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.212E-07 | 3.702E-05 |
|---|
| regulation of cell-matrix adhesion | GO:0001952 |  | 1.542E-07 | 4.186E-05 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 1.873E-07 | 4.575E-05 |
|---|
| motor axon guidance | GO:0008045 |  | 1.873E-07 | 4.159E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.641E-10 | 6.355E-07 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.739E-10 | 3.294E-07 |
|---|
| interphase | GO:0051325 |  | 3.606E-10 | 2.892E-07 |
|---|
| response to UV | GO:0009411 |  | 8.981E-10 | 5.402E-07 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 3.982E-08 | 1.916E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 8.107E-08 | 3.251E-05 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 9.835E-08 | 3.38E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.285E-07 | 3.864E-05 |
|---|
| motor axon guidance | GO:0008045 |  | 1.452E-07 | 3.883E-05 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.655E-07 | 3.981E-05 |
|---|
| regulation of cell-matrix adhesion | GO:0001952 |  | 2.104E-07 | 4.602E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.001E-09 | 2.302E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.052E-09 | 1.21E-06 |
|---|
| interphase | GO:0051325 |  | 1.26E-09 | 9.657E-07 |
|---|
| response to light stimulus | GO:0009416 |  | 4.819E-08 | 2.771E-05 |
|---|
| response to UV | GO:0009411 |  | 5.261E-08 | 2.42E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 6.676E-08 | 2.559E-05 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 1.87E-07 | 6.145E-05 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.189E-07 | 6.293E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.189E-07 | 5.594E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.523E-07 | 5.802E-05 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 2.523E-07 | 5.274E-05 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| T cell homeostasis | GO:0043029 |  | 1.431E-07 | 2.637E-04 |
|---|
| lymphocyte homeostasis | GO:0002260 |  | 4.248E-07 | 3.915E-04 |
|---|
| positive regulation of multicellular organism growth | GO:0040018 |  | 9.911E-07 | 6.089E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.187E-06 | 5.47E-04 |
|---|
| response to light stimulus | GO:0009416 |  | 1.335E-06 | 4.921E-04 |
|---|
| regulation of cell-substrate adhesion | GO:0010810 |  | 1.424E-06 | 4.375E-04 |
|---|
| response to hypoxia | GO:0001666 |  | 1.497E-06 | 3.943E-04 |
|---|
| response to oxygen levels | GO:0070482 |  | 1.676E-06 | 3.861E-04 |
|---|
| regulation of cell adhesion | GO:0030155 |  | 1.871E-06 | 3.832E-04 |
|---|
| leukocyte homeostasis | GO:0001776 |  | 2.691E-06 | 4.96E-04 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 2.848E-06 | 4.771E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.001E-09 | 2.302E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.052E-09 | 1.21E-06 |
|---|
| interphase | GO:0051325 |  | 1.26E-09 | 9.657E-07 |
|---|
| response to light stimulus | GO:0009416 |  | 4.819E-08 | 2.771E-05 |
|---|
| response to UV | GO:0009411 |  | 5.261E-08 | 2.42E-05 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 6.676E-08 | 2.559E-05 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 1.87E-07 | 6.145E-05 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.189E-07 | 6.293E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.189E-07 | 5.594E-05 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.523E-07 | 5.802E-05 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 2.523E-07 | 5.274E-05 |
|---|



