Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6367
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7572 | 3.311e-03 | 5.300e-04 | 7.619e-01 | 1.337e-06 |
|---|
| Loi | 0.2291 | 8.210e-02 | 1.185e-02 | 4.326e-01 | 4.208e-04 |
|---|
| Schmidt | 0.6720 | 0.000e+00 | 0.000e+00 | 4.642e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7906 | 0.000e+00 | 0.000e+00 | 3.495e-03 | 0.000e+00 |
|---|
| Wang | 0.2603 | 2.849e-03 | 4.536e-02 | 4.035e-01 | 5.214e-05 |
|---|
Expression data for subnetwork 6367 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
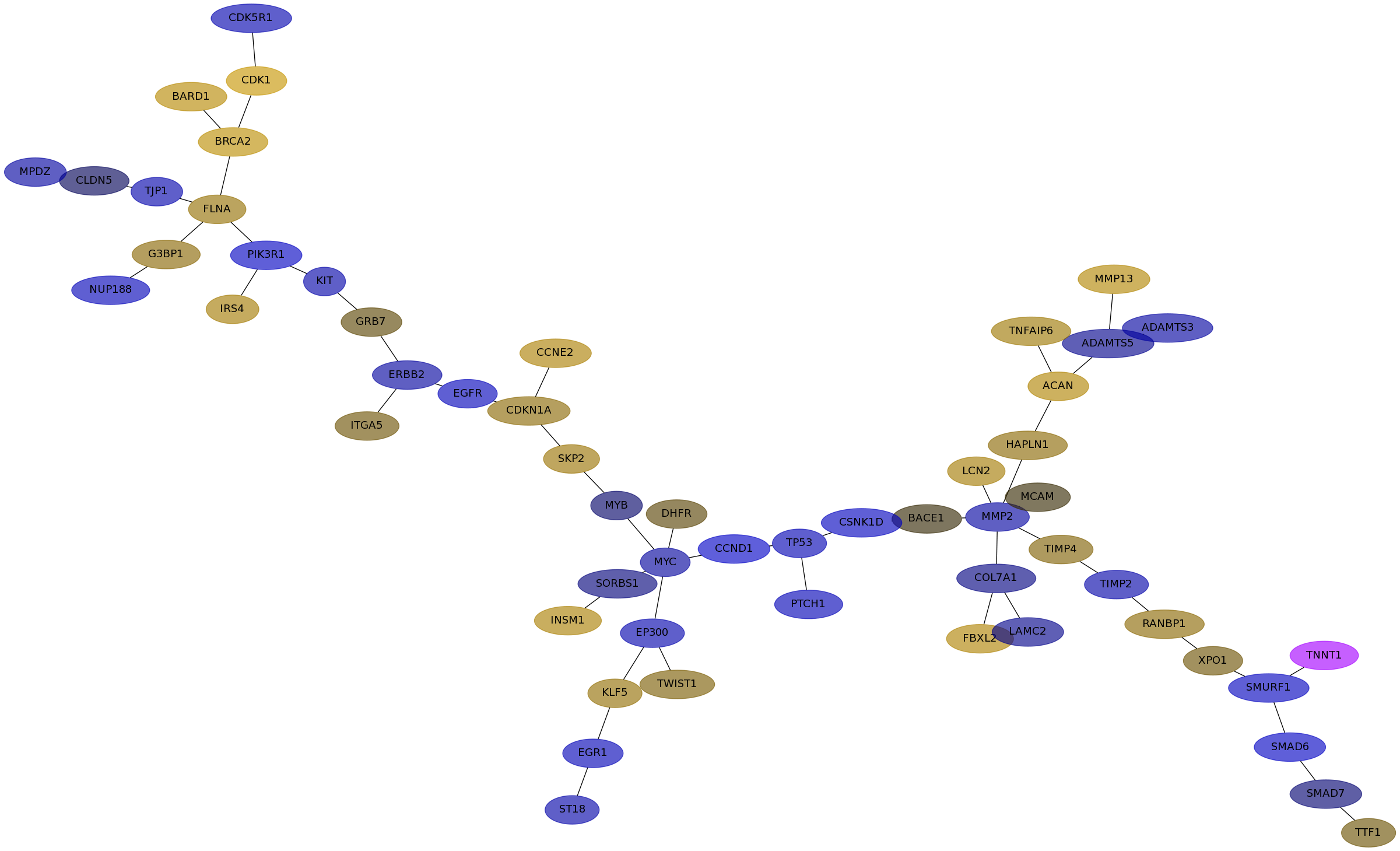
Score for each gene in subnetwork 6367 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| CCNE2 |   | 11 | 148 | 412 | 407 | 0.136 | -0.178 | 0.241 | undef | -0.055 |
|---|
| CLDN5 |   | 2 | 743 | 806 | 806 | -0.019 | 0.030 | 0.223 | -0.002 | 0.037 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| NUP188 |   | 8 | 222 | 1116 | 1079 | -0.187 | 0.094 | 0.058 | 0.161 | -0.070 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| LCN2 |   | 1 | 1195 | 2293 | 2293 | 0.113 | 0.145 | -0.098 | 0.074 | -0.284 |
|---|
| BACE1 |   | 1 | 1195 | 2293 | 2293 | 0.007 | 0.162 | 0.210 | undef | 0.090 |
|---|
| TIMP4 |   | 2 | 743 | 366 | 395 | 0.050 | 0.044 | 0.193 | -0.037 | -0.153 |
|---|
| SMAD7 |   | 5 | 360 | 1034 | 1004 | -0.035 | 0.131 | 0.169 | 0.061 | 0.147 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| TJP1 |   | 8 | 222 | 318 | 325 | -0.139 | 0.102 | 0.099 | 0.319 | 0.049 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| TTF1 |   | 2 | 743 | 1793 | 1779 | 0.031 | 0.244 | 0.114 | 0.231 | 0.008 |
|---|
| HAPLN1 |   | 1 | 1195 | 2293 | 2293 | 0.064 | 0.036 | 0.176 | undef | 0.226 |
|---|
| CCND1 |   | 26 | 52 | 1 | 24 | -0.258 | 0.074 | 0.117 | undef | 0.003 |
|---|
| MCAM |   | 1 | 1195 | 2293 | 2293 | 0.008 | 0.078 | 0.230 | -0.086 | 0.109 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| TNNT1 |   | 3 | 557 | 1914 | 1877 | undef | undef | undef | 0.145 | undef |
|---|
| RANBP1 |   | 2 | 743 | 842 | 854 | 0.063 | -0.066 | 0.199 | -0.107 | -0.048 |
|---|
| MMP2 |   | 3 | 557 | 1190 | 1182 | -0.105 | 0.186 | -0.024 | undef | 0.162 |
|---|
| ST18 |   | 16 | 104 | 236 | 227 | -0.131 | 0.028 | -0.088 | 0.186 | 0.151 |
|---|
| MPDZ |   | 3 | 557 | 1846 | 1817 | -0.110 | 0.062 | 0.231 | 0.291 | 0.167 |
|---|
| INSM1 |   | 23 | 66 | 570 | 545 | 0.135 | -0.184 | -0.188 | 0.237 | -0.032 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| FBXL2 |   | 2 | 743 | 1222 | 1215 | 0.151 | 0.089 | 0.089 | undef | -0.116 |
|---|
| MMP13 |   | 1 | 1195 | 2293 | 2293 | 0.162 | 0.112 | -0.115 | undef | 0.177 |
|---|
| SMAD6 |   | 2 | 743 | 1879 | 1866 | -0.241 | 0.053 | 0.266 | -0.043 | 0.070 |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| MYB |   | 28 | 47 | 366 | 344 | -0.028 | 0.075 | 0.002 | 0.065 | -0.012 |
|---|
| TNFAIP6 |   | 3 | 557 | 1735 | 1718 | 0.102 | 0.234 | 0.077 | -0.002 | -0.073 |
|---|
| TIMP2 |   | 2 | 743 | 1190 | 1189 | -0.126 | 0.212 | 0.021 | 0.065 | 0.137 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| SKP2 |   | 29 | 45 | 1 | 23 | 0.092 | -0.043 | 0.237 | undef | -0.009 |
|---|
| ITGA5 |   | 26 | 52 | 179 | 177 | 0.031 | 0.192 | 0.125 | 0.133 | 0.202 |
|---|
| PTCH1 |   | 5 | 360 | 1190 | 1157 | -0.184 | 0.047 | 0.121 | -0.055 | 0.004 |
|---|
| COL7A1 |   | 3 | 557 | 1222 | 1210 | -0.052 | 0.240 | 0.233 | -0.019 | 0.022 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| CDK5R1 |   | 17 | 95 | 552 | 521 | -0.150 | 0.038 | -0.096 | 0.121 | 0.091 |
|---|
| ADAMTS5 |   | 2 | 743 | 412 | 439 | -0.066 | 0.021 | 0.211 | 0.180 | 0.030 |
|---|
| XPO1 |   | 2 | 743 | 1294 | 1286 | 0.031 | -0.140 | 0.179 | 0.061 | -0.082 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| ADAMTS3 |   | 1 | 1195 | 2293 | 2293 | -0.106 | -0.078 | 0.297 | undef | 0.024 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| ACAN |   | 5 | 360 | 412 | 416 | 0.156 | 0.132 | -0.024 | -0.171 | -0.083 |
|---|
| DHFR |   | 4 | 440 | 179 | 209 | 0.019 | 0.029 | 0.232 | 0.129 | -0.050 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| SMURF1 |   | 5 | 360 | 682 | 671 | -0.222 | 0.236 | -0.049 | undef | -0.006 |
|---|
| G3BP1 |   | 7 | 256 | 1116 | 1082 | 0.060 | -0.077 | -0.017 | 0.155 | -0.004 |
|---|
| LAMC2 |   | 4 | 440 | 179 | 209 | -0.063 | 0.307 | -0.060 | -0.148 | 0.055 |
|---|
| CSNK1D |   | 6 | 301 | 614 | 602 | -0.222 | -0.044 | 0.170 | -0.088 | 0.013 |
|---|
GO Enrichment output for subnetwork 6367 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycine biosynthetic process | GO:0006545 |  | 1.217E-09 | 2.973E-06 |
|---|
| response to UV | GO:0009411 |  | 9.021E-08 | 1.102E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.122E-07 | 9.135E-05 |
|---|
| serine family amino acid biosynthetic process | GO:0009070 |  | 2.369E-07 | 1.447E-04 |
|---|
| negative regulation of DNA metabolic process | GO:0051053 |  | 2.795E-07 | 1.365E-04 |
|---|
| glycine metabolic process | GO:0006544 |  | 7.153E-07 | 2.912E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.019E-06 | 3.555E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.019E-06 | 3.11E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.139E-06 | 3.092E-04 |
|---|
| collagen catabolic process | GO:0030574 |  | 1.386E-06 | 3.386E-04 |
|---|
| regulation of helicase activity | GO:0051095 |  | 2.159E-06 | 4.796E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycine biosynthetic process | GO:0006545 |  | 1.601E-09 | 3.852E-06 |
|---|
| response to UV | GO:0009411 |  | 1.069E-07 | 1.286E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.667E-07 | 1.337E-04 |
|---|
| serine family amino acid biosynthetic process | GO:0009070 |  | 2.229E-07 | 1.341E-04 |
|---|
| negative regulation of DNA metabolic process | GO:0051053 |  | 3.905E-07 | 1.879E-04 |
|---|
| glycine metabolic process | GO:0006544 |  | 7.322E-07 | 2.936E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.127E-06 | 3.872E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.42E-06 | 4.271E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.588E-06 | 4.245E-04 |
|---|
| collagen catabolic process | GO:0030574 |  | 1.817E-06 | 4.372E-04 |
|---|
| regulation of helicase activity | GO:0051095 |  | 2.654E-06 | 5.804E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to UV | GO:0009411 |  | 2.96E-07 | 6.807E-04 |
|---|
| serine family amino acid biosynthetic process | GO:0009070 |  | 3.777E-07 | 4.344E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.555E-07 | 4.259E-04 |
|---|
| glycine metabolic process | GO:0006544 |  | 1.13E-06 | 6.498E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 3.316E-06 | 1.525E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.771E-06 | 1.446E-03 |
|---|
| collagen catabolic process | GO:0030574 |  | 5.321E-06 | 1.748E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.44E-06 | 1.564E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.036E-06 | 1.798E-03 |
|---|
| negative regulation of DNA replication | GO:0008156 |  | 9.593E-06 | 2.206E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.121E-05 | 2.344E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell junction assembly | GO:0034329 |  | 1.275E-06 | 2.349E-03 |
|---|
| cell junction organization | GO:0034330 |  | 3.848E-06 | 3.546E-03 |
|---|
| cell-substrate junction assembly | GO:0007044 |  | 1.413E-05 | 8.679E-03 |
|---|
| regulation of protein catabolic process | GO:0042176 |  | 2.336E-05 | 0.01076315 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 4.226E-05 | 0.0155758 |
|---|
| regulation of catabolic process | GO:0009894 |  | 8.573E-05 | 0.0263323 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 9.342E-05 | 0.02459501 |
|---|
| BMP signaling pathway | GO:0030509 |  | 9.342E-05 | 0.02152063 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.011E-04 | 0.02070312 |
|---|
| negative regulation of protein transport | GO:0051224 |  | 1.507E-04 | 0.02776901 |
|---|
| T cell differentiation | GO:0030217 |  | 2.269E-04 | 0.03801737 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to UV | GO:0009411 |  | 2.96E-07 | 6.807E-04 |
|---|
| serine family amino acid biosynthetic process | GO:0009070 |  | 3.777E-07 | 4.344E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.555E-07 | 4.259E-04 |
|---|
| glycine metabolic process | GO:0006544 |  | 1.13E-06 | 6.498E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 3.316E-06 | 1.525E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.771E-06 | 1.446E-03 |
|---|
| collagen catabolic process | GO:0030574 |  | 5.321E-06 | 1.748E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.44E-06 | 1.564E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.036E-06 | 1.798E-03 |
|---|
| negative regulation of DNA replication | GO:0008156 |  | 9.593E-06 | 2.206E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.121E-05 | 2.344E-03 |
|---|



