Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6334
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7575 | 3.294e-03 | 5.260e-04 | 7.611e-01 | 1.319e-06 |
|---|
| Loi | 0.2293 | 8.189e-02 | 1.179e-02 | 4.322e-01 | 4.174e-04 |
|---|
| Schmidt | 0.6717 | 0.000e+00 | 0.000e+00 | 4.674e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.483e-03 | 0.000e+00 |
|---|
| Wang | 0.2602 | 2.872e-03 | 4.556e-02 | 4.044e-01 | 5.291e-05 |
|---|
Expression data for subnetwork 6334 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
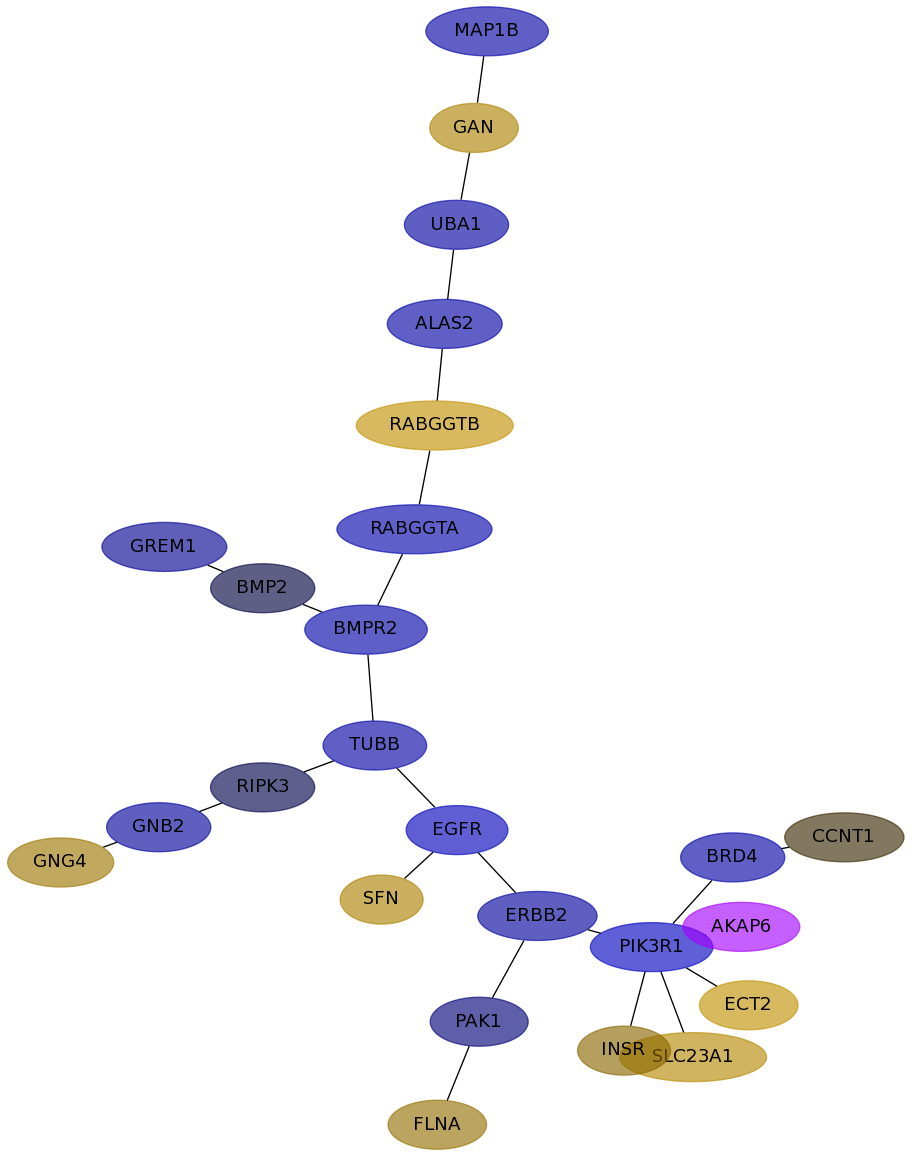
Score for each gene in subnetwork 6334 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| PAK1 |   | 3 | 557 | 682 | 685 | -0.040 | -0.122 | -0.143 | 0.075 | 0.006 |
|---|
| GREM1 |   | 9 | 196 | 525 | 508 | -0.073 | 0.200 | 0.028 | 0.225 | 0.261 |
|---|
| BRD4 |   | 11 | 148 | 1047 | 1003 | -0.119 | 0.095 | -0.075 | 0.081 | -0.044 |
|---|
| BMPR2 |   | 5 | 360 | 1323 | 1292 | -0.132 | 0.081 | -0.029 | 0.356 | -0.021 |
|---|
| UBA1 |   | 1 | 1195 | 2260 | 2261 | -0.109 | 0.204 | -0.166 | 0.072 | -0.169 |
|---|
| SLC23A1 |   | 2 | 743 | 457 | 481 | 0.179 | 0.104 | undef | 0.052 | undef |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| RIPK3 |   | 19 | 85 | 179 | 186 | -0.014 | 0.040 | undef | 0.173 | undef |
|---|
| ECT2 |   | 1 | 1195 | 2260 | 2261 | 0.223 | -0.045 | 0.040 | 0.086 | -0.058 |
|---|
| GNG4 |   | 5 | 360 | 1077 | 1057 | 0.095 | -0.139 | -0.169 | 0.129 | 0.106 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| ALAS2 |   | 1 | 1195 | 2260 | 2261 | -0.113 | 0.057 | undef | -0.016 | undef |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| GNB2 |   | 11 | 148 | 525 | 505 | -0.091 | 0.082 | -0.062 | 0.208 | -0.062 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| RABGGTA |   | 1 | 1195 | 2260 | 2261 | -0.136 | 0.170 | -0.021 | 0.096 | 0.031 |
|---|
| AKAP6 |   | 3 | 557 | 882 | 871 | undef | 0.116 | -0.125 | 0.026 | 0.153 |
|---|
| BMP2 |   | 11 | 148 | 525 | 505 | -0.011 | 0.097 | 0.148 | -0.114 | -0.069 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| CCNT1 |   | 6 | 301 | 478 | 473 | 0.008 | -0.028 | 0.162 | 0.007 | -0.213 |
|---|
| GAN |   | 1 | 1195 | 2260 | 2261 | 0.145 | 0.250 | -0.086 | undef | -0.026 |
|---|
| RABGGTB |   | 1 | 1195 | 2260 | 2261 | 0.233 | -0.019 | 0.166 | 0.053 | 0.153 |
|---|
| SFN |   | 25 | 59 | 478 | 454 | 0.141 | 0.270 | 0.023 | -0.102 | 0.101 |
|---|
| TUBB |   | 5 | 360 | 614 | 612 | -0.118 | 0.170 | 0.200 | undef | -0.045 |
|---|
GO Enrichment output for subnetwork 6334 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 5.755E-11 | 1.406E-07 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 2.4E-09 | 2.932E-06 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 5.642E-09 | 4.594E-06 |
|---|
| cell killing | GO:0001906 |  | 1.549E-08 | 9.461E-06 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.43E-07 | 6.986E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 3.561E-07 | 1.45E-04 |
|---|
| spindle organization | GO:0007051 |  | 3.984E-07 | 1.39E-04 |
|---|
| receptor clustering | GO:0043113 |  | 5.334E-07 | 1.629E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 5.967E-07 | 1.62E-04 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 9.344E-07 | 2.283E-04 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 2.296E-06 | 5.099E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 8.676E-11 | 2.088E-07 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 3.616E-09 | 4.35E-06 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 8.496E-09 | 6.814E-06 |
|---|
| cell killing | GO:0001906 |  | 2.332E-08 | 1.403E-05 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.703E-07 | 8.193E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 4.849E-07 | 1.944E-04 |
|---|
| spindle organization | GO:0007051 |  | 5.981E-07 | 2.056E-04 |
|---|
| receptor clustering | GO:0043113 |  | 7.262E-07 | 2.184E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 8.211E-07 | 2.195E-04 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.537E-06 | 3.697E-04 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 3.123E-06 | 6.832E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 1.583E-10 | 3.641E-07 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 3.961E-09 | 4.555E-06 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 1.034E-08 | 7.926E-06 |
|---|
| cell killing | GO:0001906 |  | 3.119E-08 | 1.793E-05 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 8.043E-08 | 3.7E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 7.696E-07 | 2.95E-04 |
|---|
| spindle organization | GO:0007051 |  | 8.299E-07 | 2.727E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.006E-06 | 2.893E-04 |
|---|
| receptor clustering | GO:0043113 |  | 1.152E-06 | 2.945E-04 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 1.894E-06 | 4.355E-04 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 2.008E-06 | 4.199E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 8.568E-09 | 1.579E-05 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.604E-07 | 1.478E-04 |
|---|
| receptor clustering | GO:0043113 |  | 1.78E-07 | 1.093E-04 |
|---|
| regulation of MAP kinase activity | GO:0043405 |  | 7.302E-07 | 3.365E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.061E-06 | 7.597E-04 |
|---|
| neuromuscular junction development | GO:0007528 |  | 2.117E-06 | 6.503E-04 |
|---|
| positive regulation of protein kinase activity | GO:0045860 |  | 2.75E-06 | 7.241E-04 |
|---|
| signal complex assembly | GO:0007172 |  | 2.906E-06 | 6.695E-04 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 2.906E-06 | 5.951E-04 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 3.083E-06 | 5.682E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 5.02E-06 | 8.41E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 1.583E-10 | 3.641E-07 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 3.961E-09 | 4.555E-06 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 1.034E-08 | 7.926E-06 |
|---|
| cell killing | GO:0001906 |  | 3.119E-08 | 1.793E-05 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 8.043E-08 | 3.7E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 7.696E-07 | 2.95E-04 |
|---|
| spindle organization | GO:0007051 |  | 8.299E-07 | 2.727E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.006E-06 | 2.893E-04 |
|---|
| receptor clustering | GO:0043113 |  | 1.152E-06 | 2.945E-04 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 1.894E-06 | 4.355E-04 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 2.008E-06 | 4.199E-04 |
|---|



