Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6313
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7579 | 3.272e-03 | 5.220e-04 | 7.601e-01 | 1.298e-06 |
|---|
| Loi | 0.2292 | 8.205e-02 | 1.183e-02 | 4.325e-01 | 4.200e-04 |
|---|
| Schmidt | 0.6716 | 0.000e+00 | 0.000e+00 | 4.687e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.483e-03 | 0.000e+00 |
|---|
| Wang | 0.2601 | 2.890e-03 | 4.572e-02 | 4.051e-01 | 5.352e-05 |
|---|
Expression data for subnetwork 6313 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
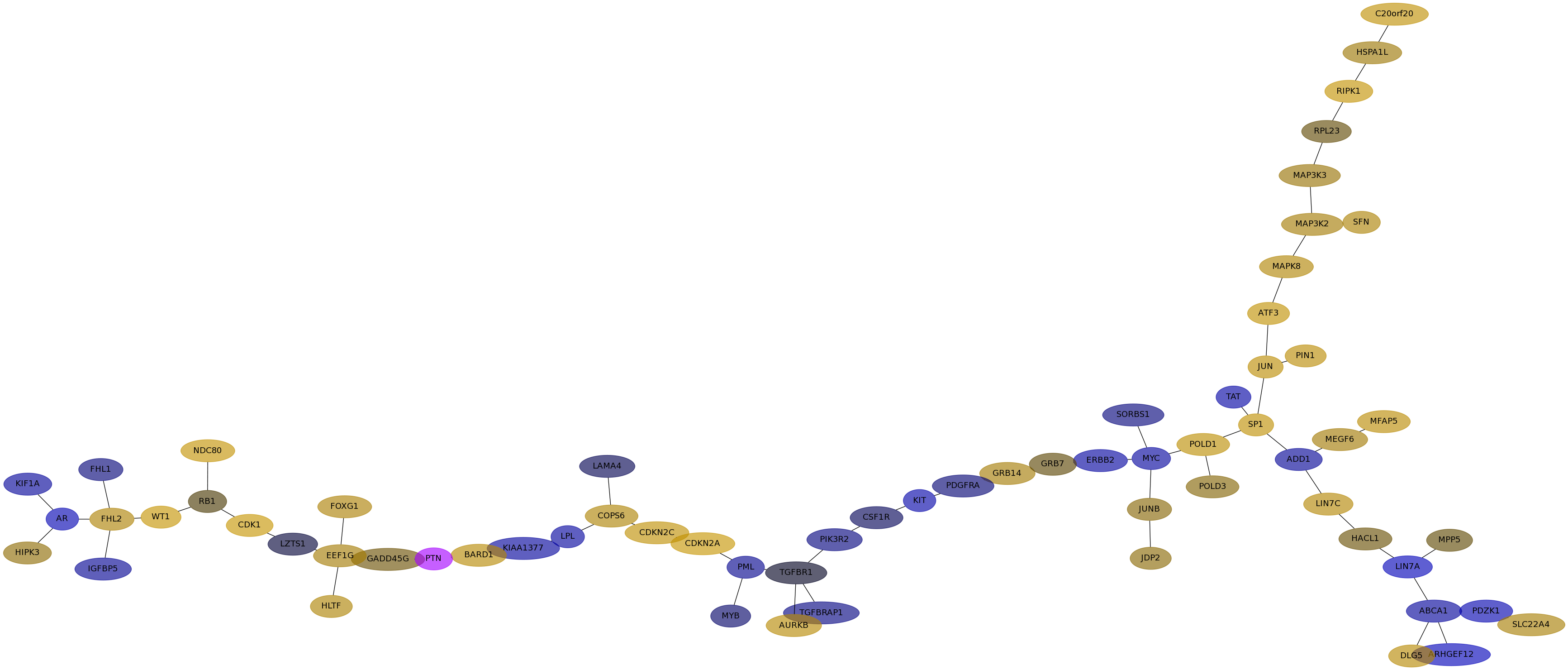
Score for each gene in subnetwork 6313 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| MAPK8 |   | 8 | 222 | 696 | 677 | 0.153 | 0.200 | 0.249 | -0.066 | 0.138 |
|---|
| HACL1 |   | 4 | 440 | 590 | 585 | 0.028 | -0.071 | undef | -0.278 | undef |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| ADD1 |   | 8 | 222 | 366 | 373 | -0.085 | 0.116 | 0.035 | undef | -0.098 |
|---|
| MFAP5 |   | 1 | 1195 | 2215 | 2217 | 0.195 | 0.036 | 0.040 | 0.071 | 0.142 |
|---|
| NDC80 |   | 2 | 743 | 525 | 547 | 0.246 | -0.229 | 0.060 | -0.005 | 0.044 |
|---|
| RIPK1 |   | 1 | 1195 | 2215 | 2217 | 0.249 | 0.088 | -0.021 | 0.020 | -0.035 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| MPP5 |   | 1 | 1195 | 2215 | 2217 | 0.022 | 0.005 | -0.090 | undef | 0.044 |
|---|
| HLTF |   | 2 | 743 | 1644 | 1630 | 0.145 | 0.070 | 0.114 | 0.209 | -0.006 |
|---|
| EEF1G |   | 8 | 222 | 882 | 869 | 0.116 | -0.224 | 0.131 | 0.086 | 0.082 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| PDGFRA |   | 10 | 167 | 141 | 146 | -0.035 | 0.008 | 0.118 | 0.107 | 0.119 |
|---|
| POLD1 |   | 1 | 1195 | 2215 | 2217 | 0.203 | -0.129 | -0.006 | -0.111 | -0.167 |
|---|
| CDKN2C |   | 16 | 104 | 236 | 227 | 0.216 | -0.106 | 0.202 | -0.024 | -0.081 |
|---|
| DLG5 |   | 1 | 1195 | 2215 | 2217 | 0.180 | 0.106 | 0.195 | 0.138 | 0.009 |
|---|
| MAP3K2 |   | 2 | 743 | 2215 | 2193 | 0.115 | 0.087 | -0.066 | -0.070 | -0.117 |
|---|
| PIN1 |   | 24 | 62 | 179 | 181 | 0.194 | 0.040 | -0.040 | 0.065 | -0.048 |
|---|
| PIK3R2 |   | 4 | 440 | 141 | 165 | -0.047 | 0.015 | -0.147 | undef | -0.135 |
|---|
| LIN7A |   | 6 | 301 | 366 | 377 | -0.189 | -0.074 | 0.108 | 0.065 | -0.146 |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| TGFBRAP1 |   | 2 | 743 | 2003 | 1980 | -0.051 | 0.018 | -0.088 | -0.094 | -0.105 |
|---|
| MEGF6 |   | 1 | 1195 | 2215 | 2217 | 0.110 | 0.197 | 0.067 | -0.045 | 0.083 |
|---|
| TAT |   | 11 | 148 | 83 | 97 | -0.113 | 0.030 | -0.182 | 0.037 | -0.074 |
|---|
| LAMA4 |   | 13 | 124 | 696 | 670 | -0.016 | 0.138 | 0.209 | 0.172 | 0.343 |
|---|
| MAP3K3 |   | 4 | 440 | 1364 | 1330 | 0.085 | 0.046 | 0.028 | -0.035 | 0.094 |
|---|
| ATF3 |   | 17 | 95 | 179 | 189 | 0.225 | -0.021 | 0.000 | undef | -0.039 |
|---|
| AURKB |   | 4 | 440 | 83 | 112 | 0.179 | -0.189 | 0.086 | -0.058 | -0.028 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| HSPA1L |   | 1 | 1195 | 2215 | 2217 | 0.097 | -0.104 | 0.033 | 0.077 | 0.095 |
|---|
| KIF1A |   | 2 | 743 | 141 | 197 | -0.089 | -0.067 | 0.178 | 0.007 | -0.047 |
|---|
| LPL |   | 13 | 124 | 236 | 230 | -0.102 | -0.062 | 0.292 | 0.087 | -0.105 |
|---|
| COPS6 |   | 16 | 104 | 1 | 31 | 0.147 | -0.068 | 0.128 | 0.216 | 0.065 |
|---|
| ABCA1 |   | 1 | 1195 | 2215 | 2217 | -0.093 | 0.037 | 0.070 | -0.001 | 0.043 |
|---|
| LZTS1 |   | 3 | 557 | 2147 | 2108 | -0.008 | -0.089 | 0.043 | 0.147 | 0.063 |
|---|
| PTN |   | 24 | 62 | 236 | 225 | undef | 0.022 | 0.000 | 0.054 | -0.092 |
|---|
| GRB14 |   | 24 | 62 | 179 | 181 | 0.113 | -0.129 | 0.143 | -0.098 | 0.138 |
|---|
| JDP2 |   | 4 | 440 | 2053 | 2008 | 0.062 | 0.045 | undef | undef | undef |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| TGFBR1 |   | 4 | 440 | 1793 | 1755 | -0.004 | 0.036 | -0.078 | 0.181 | undef |
|---|
| KIAA1377 |   | 18 | 86 | 141 | 136 | -0.095 | 0.116 | undef | 0.198 | undef |
|---|
| FOXG1 |   | 6 | 301 | 727 | 716 | 0.136 | -0.156 | -0.126 | -0.013 | -0.068 |
|---|
| C20orf20 |   | 2 | 743 | 906 | 911 | 0.221 | -0.160 | 0.161 | 0.018 | 0.024 |
|---|
| JUN |   | 26 | 52 | 179 | 177 | 0.200 | 0.051 | 0.220 | undef | 0.200 |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| RPL23 |   | 2 | 743 | 2215 | 2193 | 0.024 | -0.022 | 0.164 | 0.086 | -0.032 |
|---|
| PDZK1 |   | 1 | 1195 | 2215 | 2217 | -0.150 | 0.145 | 0.209 | -0.184 | -0.051 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| JUNB |   | 20 | 81 | 179 | 185 | 0.057 | 0.086 | 0.065 | 0.001 | -0.015 |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| MYB |   | 28 | 47 | 366 | 344 | -0.028 | 0.075 | 0.002 | 0.065 | -0.012 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| FHL2 |   | 21 | 73 | 658 | 630 | 0.143 | 0.239 | -0.033 | 0.107 | 0.294 |
|---|
| ARHGEF12 |   | 2 | 743 | 1316 | 1312 | -0.190 | 0.167 | -0.018 | 0.206 | 0.126 |
|---|
| POLD3 |   | 2 | 743 | 1077 | 1077 | 0.053 | -0.072 | 0.193 | 0.089 | 0.042 |
|---|
| LIN7C |   | 10 | 167 | 590 | 566 | 0.166 | 0.088 | 0.118 | -0.018 | 0.094 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| PML |   | 6 | 301 | 478 | 473 | -0.068 | -0.010 | -0.007 | -0.153 | -0.070 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| FHL1 |   | 14 | 117 | 614 | 591 | -0.041 | 0.104 | 0.301 | undef | 0.266 |
|---|
| HIPK3 |   | 9 | 196 | 1461 | 1417 | 0.069 | 0.005 | -0.027 | undef | 0.175 |
|---|
| SLC22A4 |   | 2 | 743 | 2197 | 2176 | 0.124 | 0.045 | 0.023 | 0.072 | 0.020 |
|---|
| SFN |   | 25 | 59 | 478 | 454 | 0.141 | 0.270 | 0.023 | -0.102 | 0.101 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| CSF1R |   | 1 | 1195 | 2215 | 2217 | -0.019 | -0.073 | -0.072 | -0.081 | -0.047 |
|---|
GO Enrichment output for subnetwork 6313 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.382E-06 | 3.376E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.505E-06 | 1.839E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.092E-06 | 1.703E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 2.829E-06 | 1.728E-03 |
|---|
| activation of pro-apoptotic gene products | GO:0008633 |  | 5.11E-06 | 2.497E-03 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 6.108E-06 | 2.487E-03 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 6.904E-06 | 2.409E-03 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 8.526E-06 | 2.604E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 1.032E-05 | 2.801E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.836E-05 | 4.485E-03 |
|---|
| negative regulation of cell growth | GO:0030308 |  | 2.297E-05 | 5.102E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.68E-06 | 4.041E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.834E-06 | 2.206E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.572E-06 | 2.063E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 3.654E-06 | 2.198E-03 |
|---|
| activation of pro-apoptotic gene products | GO:0008633 |  | 6.595E-06 | 3.174E-03 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 7.881E-06 | 3.16E-03 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 8.388E-06 | 2.883E-03 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.1E-05 | 3.307E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 1.253E-05 | 3.35E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 1.734E-05 | 4.171E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.82E-05 | 3.982E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.044E-06 | 7.002E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 3.376E-06 | 3.882E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 4.551E-06 | 3.489E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 6.832E-06 | 3.928E-03 |
|---|
| activation of pro-apoptotic gene products | GO:0008633 |  | 1.314E-05 | 6.046E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.612E-05 | 6.179E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 1.901E-05 | 6.245E-03 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 1.901E-05 | 5.464E-03 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.923E-05 | 4.915E-03 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 2.718E-05 | 6.251E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 3.336E-05 | 6.975E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 3.157E-06 | 5.818E-03 |
|---|
| regulation of cell growth | GO:0001558 |  | 3.813E-06 | 3.514E-03 |
|---|
| regulation of cell size | GO:0008361 |  | 6.598E-06 | 4.054E-03 |
|---|
| negative regulation of cell growth | GO:0030308 |  | 6.499E-05 | 0.02994261 |
|---|
| negative regulation of cell size | GO:0045792 |  | 7.357E-05 | 0.02711894 |
|---|
| nucleotide-excision repair. DNA gap filling | GO:0006297 |  | 8.68E-05 | 0.0266634 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.163E-04 | 0.03063091 |
|---|
| negative regulation of growth | GO:0045926 |  | 1.163E-04 | 0.02680205 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.294E-04 | 0.026491 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 1.585E-04 | 0.02921812 |
|---|
| triglyceride metabolic process | GO:0006641 |  | 1.675E-04 | 0.02806089 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.044E-06 | 7.002E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 3.376E-06 | 3.882E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 4.551E-06 | 3.489E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 6.832E-06 | 3.928E-03 |
|---|
| activation of pro-apoptotic gene products | GO:0008633 |  | 1.314E-05 | 6.046E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.612E-05 | 6.179E-03 |
|---|
| DNA synthesis during DNA repair | GO:0000731 |  | 1.901E-05 | 6.245E-03 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 1.901E-05 | 5.464E-03 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.923E-05 | 4.915E-03 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 2.718E-05 | 6.251E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 3.336E-05 | 6.975E-03 |
|---|



