Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6252
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7594 | 3.181e-03 | 5.030e-04 | 7.557e-01 | 1.209e-06 |
|---|
| Loi | 0.2290 | 8.224e-02 | 1.188e-02 | 4.330e-01 | 4.231e-04 |
|---|
| Schmidt | 0.6710 | 0.000e+00 | 0.000e+00 | 4.752e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.479e-03 | 0.000e+00 |
|---|
| Wang | 0.2594 | 2.981e-03 | 4.652e-02 | 4.087e-01 | 5.667e-05 |
|---|
Expression data for subnetwork 6252 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
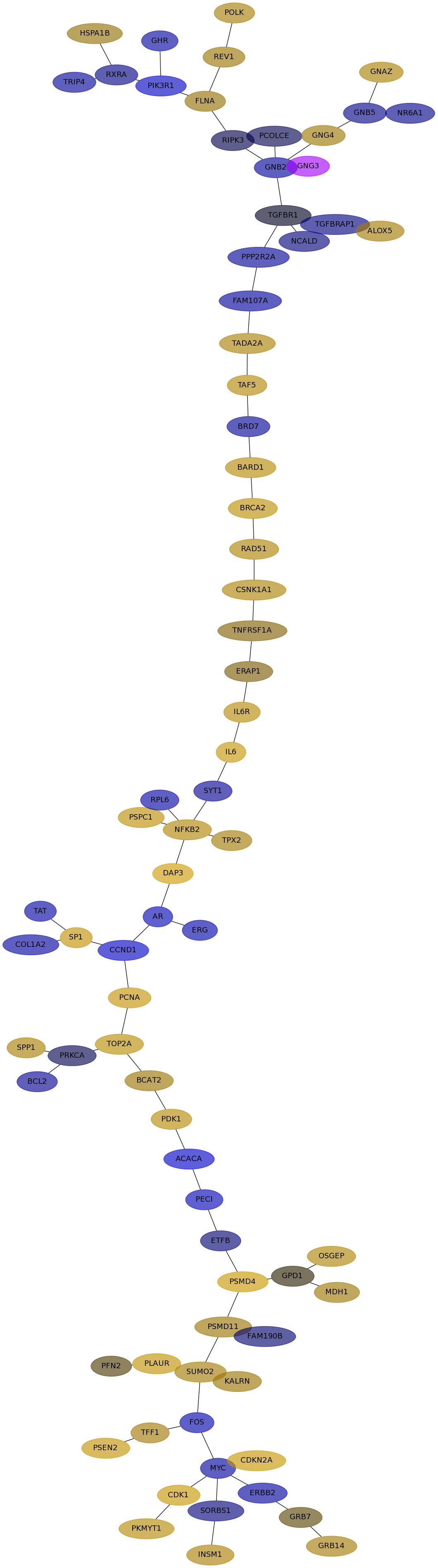
Score for each gene in subnetwork 6252 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| GNG3 |   | 1 | 1195 | 2003 | 2010 | undef | -0.034 | 0.230 | 0.181 | -0.036 |
|---|
| SUMO2 |   | 22 | 69 | 366 | 345 | 0.117 | 0.062 | 0.294 | 0.141 | -0.096 |
|---|
| RXRA |   | 7 | 256 | 457 | 451 | -0.055 | 0.134 | -0.027 | 0.022 | -0.096 |
|---|
| TPX2 |   | 5 | 360 | 412 | 416 | 0.114 | -0.121 | -0.018 | undef | -0.056 |
|---|
| PPP2R2A |   | 1 | 1195 | 2003 | 2010 | -0.084 | 0.001 | 0.123 | 0.067 | -0.167 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| GNB2 |   | 11 | 148 | 525 | 505 | -0.091 | 0.082 | -0.062 | 0.208 | -0.062 |
|---|
| BCAT2 |   | 3 | 557 | 1510 | 1486 | 0.091 | -0.103 | -0.096 | -0.128 | -0.030 |
|---|
| ALOX5 |   | 2 | 743 | 1681 | 1665 | 0.115 | 0.016 | 0.137 | -0.198 | 0.037 |
|---|
| CCND1 |   | 26 | 52 | 1 | 24 | -0.258 | 0.074 | 0.117 | undef | 0.003 |
|---|
| PCNA |   | 15 | 111 | 1 | 32 | 0.243 | -0.158 | 0.027 | -0.099 | 0.003 |
|---|
| GNAZ |   | 15 | 111 | 478 | 457 | 0.134 | -0.116 | 0.308 | 0.267 | 0.011 |
|---|
| FAM107A |   | 1 | 1195 | 2003 | 2010 | -0.102 | -0.043 | 0.144 | -0.248 | -0.014 |
|---|
| PSEN2 |   | 14 | 117 | 614 | 591 | 0.239 | -0.068 | -0.157 | 0.295 | 0.045 |
|---|
| NFKB2 |   | 7 | 256 | 1116 | 1082 | 0.153 | -0.239 | -0.131 | undef | -0.065 |
|---|
| TGFBRAP1 |   | 2 | 743 | 2003 | 1980 | -0.051 | 0.018 | -0.088 | -0.094 | -0.105 |
|---|
| TADA2A |   | 4 | 440 | 478 | 479 | 0.135 | -0.054 | 0.204 | -0.115 | -0.127 |
|---|
| ACACA |   | 3 | 557 | 1413 | 1407 | -0.275 | -0.103 | 0.056 | undef | -0.080 |
|---|
| TAT |   | 11 | 148 | 83 | 97 | -0.113 | 0.030 | -0.182 | 0.037 | -0.074 |
|---|
| TAF5 |   | 5 | 360 | 478 | 476 | 0.166 | -0.131 | 0.125 | undef | 0.076 |
|---|
| PDK1 |   | 10 | 167 | 1 | 40 | 0.183 | -0.138 | -0.041 | -0.265 | -0.126 |
|---|
| NR6A1 |   | 1 | 1195 | 2003 | 2010 | -0.064 | 0.150 | 0.000 | -0.372 | -0.091 |
|---|
| KALRN |   | 4 | 440 | 1102 | 1080 | 0.090 | 0.136 | -0.057 | 0.202 | -0.053 |
|---|
| OSGEP |   | 1 | 1195 | 2003 | 2010 | 0.147 | 0.032 | 0.004 | -0.082 | -0.155 |
|---|
| PKMYT1 |   | 7 | 256 | 1510 | 1463 | 0.175 | -0.143 | 0.046 | 0.105 | -0.070 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| DAP3 |   | 3 | 557 | 1425 | 1409 | 0.306 | -0.088 | -0.047 | 0.166 | 0.081 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| NCALD |   | 4 | 440 | 1709 | 1672 | -0.046 | 0.078 | 0.090 | undef | 0.004 |
|---|
| TNFRSF1A |   | 8 | 222 | 412 | 413 | 0.050 | 0.094 | -0.014 | 0.214 | -0.045 |
|---|
| PCOLCE |   | 2 | 743 | 525 | 547 | -0.016 | 0.150 | 0.032 | 0.004 | 0.127 |
|---|
| GRB14 |   | 24 | 62 | 179 | 181 | 0.113 | -0.129 | 0.143 | -0.098 | 0.138 |
|---|
| TGFBR1 |   | 4 | 440 | 1793 | 1755 | -0.004 | 0.036 | -0.078 | 0.181 | undef |
|---|
| FAM190B |   | 1 | 1195 | 2003 | 2010 | -0.032 | 0.110 | 0.180 | undef | 0.140 |
|---|
| PFN2 |   | 11 | 148 | 141 | 143 | 0.014 | -0.001 | 0.286 | -0.002 | -0.056 |
|---|
| CSNK1A1 |   | 2 | 743 | 1364 | 1363 | 0.141 | 0.258 | 0.067 | undef | 0.086 |
|---|
| ETFB |   | 2 | 743 | 2003 | 1980 | -0.037 | 0.058 | 0.081 | -0.105 | -0.065 |
|---|
| GPD1 |   | 7 | 256 | 658 | 644 | 0.006 | -0.019 | 0.112 | -0.034 | 0.033 |
|---|
| TOP2A |   | 20 | 81 | 1 | 28 | 0.193 | -0.168 | 0.054 | -0.064 | 0.071 |
|---|
| SPP1 |   | 26 | 52 | 83 | 91 | 0.130 | 0.119 | 0.184 | 0.204 | 0.153 |
|---|
| GNB5 |   | 9 | 196 | 1131 | 1100 | -0.059 | 0.026 | 0.064 | 0.284 | 0.103 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| BRD7 |   | 23 | 66 | 457 | 437 | -0.090 | 0.019 | -0.188 | undef | -0.086 |
|---|
| PECI |   | 2 | 743 | 1 | 66 | -0.174 | 0.127 | 0.056 | -0.011 | 0.051 |
|---|
| PSMD11 |   | 6 | 301 | 1437 | 1408 | 0.088 | -0.090 | -0.137 | 0.060 | -0.078 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| INSM1 |   | 23 | 66 | 570 | 545 | 0.135 | -0.184 | -0.188 | 0.237 | -0.032 |
|---|
| TRIP4 |   | 1 | 1195 | 2003 | 2010 | -0.091 | 0.246 | -0.063 | -0.035 | 0.019 |
|---|
| REV1 |   | 9 | 196 | 296 | 285 | 0.090 | -0.027 | 0.165 | 0.049 | 0.039 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| ERG |   | 9 | 196 | 179 | 195 | -0.134 | -0.026 | 0.235 | -0.027 | 0.182 |
|---|
| POLK |   | 3 | 557 | 806 | 792 | 0.120 | 0.046 | undef | 0.113 | undef |
|---|
| PSPC1 |   | 6 | 301 | 991 | 964 | 0.203 | -0.289 | 0.298 | 0.148 | -0.107 |
|---|
| PLAUR |   | 5 | 360 | 696 | 680 | 0.232 | 0.083 | 0.022 | -0.030 | 0.108 |
|---|
| RIPK3 |   | 19 | 85 | 179 | 186 | -0.014 | 0.040 | undef | 0.173 | undef |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| RPL6 |   | 1 | 1195 | 2003 | 2010 | -0.117 | 0.136 | -0.032 | -0.276 | -0.105 |
|---|
| GNG4 |   | 5 | 360 | 1077 | 1057 | 0.095 | -0.139 | -0.169 | 0.129 | 0.106 |
|---|
| IL6R |   | 1 | 1195 | 2003 | 2010 | 0.179 | -0.045 | 0.002 | undef | -0.072 |
|---|
| PSMD4 |   | 11 | 148 | 296 | 282 | 0.285 | -0.243 | -0.043 | 0.087 | -0.045 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| SYT1 |   | 11 | 148 | 1 | 39 | -0.079 | 0.051 | 0.038 | 0.189 | -0.124 |
|---|
| HSPA1B |   | 2 | 743 | 1914 | 1897 | 0.075 | 0.117 | -0.061 | undef | 0.223 |
|---|
| TFF1 |   | 7 | 256 | 957 | 928 | 0.106 | 0.022 | 0.088 | -0.033 | -0.244 |
|---|
| RAD51 |   | 13 | 124 | 727 | 699 | 0.146 | -0.098 | 0.135 | -0.070 | 0.061 |
|---|
| IL6 |   | 9 | 196 | 366 | 365 | 0.257 | -0.106 | 0.226 | 0.024 | 0.035 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| ERAP1 |   | 1 | 1195 | 2003 | 2010 | 0.042 | -0.065 | 0.104 | 0.149 | -0.033 |
|---|
| MDH1 |   | 2 | 743 | 1437 | 1433 | 0.098 | -0.088 | 0.153 | -0.030 | -0.058 |
|---|
| COL1A2 |   | 7 | 256 | 552 | 542 | -0.110 | 0.207 | 0.046 | 0.235 | 0.097 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
GO Enrichment output for subnetwork 6252 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 1.015E-07 | 2.479E-04 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 1.307E-07 | 1.596E-04 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 1.662E-07 | 1.354E-04 |
|---|
| positive regulation of response to external stimulus | GO:0032103 |  | 1.739E-07 | 1.062E-04 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 2.103E-07 | 1.027E-04 |
|---|
| regulation of behavior | GO:0050795 |  | 2.601E-07 | 1.059E-04 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 3.308E-07 | 1.154E-04 |
|---|
| response to steroid hormone stimulus | GO:0048545 |  | 3.308E-07 | 1.01E-04 |
|---|
| positive regulation of phosphorylation | GO:0042327 |  | 3.93E-07 | 1.067E-04 |
|---|
| positive regulation of phosphorus metabolic process | GO:0010562 |  | 4.649E-07 | 1.136E-04 |
|---|
| regulation of ligase activity | GO:0051340 |  | 5.047E-07 | 1.121E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of locomotion | GO:0040017 |  | 1.475E-07 | 3.548E-04 |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 1.475E-07 | 1.774E-04 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 1.899E-07 | 1.523E-04 |
|---|
| positive regulation of response to external stimulus | GO:0032103 |  | 2.345E-07 | 1.41E-04 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 2.878E-07 | 1.385E-04 |
|---|
| regulation of behavior | GO:0050795 |  | 3.035E-07 | 1.217E-04 |
|---|
| response to steroid hormone stimulus | GO:0048545 |  | 5.005E-07 | 1.72E-04 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 5.464E-07 | 1.643E-04 |
|---|
| positive regulation of phosphorylation | GO:0042327 |  | 5.464E-07 | 1.461E-04 |
|---|
| positive regulation of phosphorus metabolic process | GO:0010562 |  | 6.488E-07 | 1.561E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 7.057E-07 | 1.544E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of locomotion | GO:0040017 |  | 3.1E-07 | 7.13E-04 |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 3.1E-07 | 3.565E-04 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 4.044E-07 | 3.1E-04 |
|---|
| positive regulation of response to external stimulus | GO:0032103 |  | 4.914E-07 | 2.826E-04 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 4.918E-07 | 2.262E-04 |
|---|
| regulation of behavior | GO:0050795 |  | 5.202E-07 | 1.994E-04 |
|---|
| positive regulation of phosphorylation | GO:0042327 |  | 9.143E-07 | 3.004E-04 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.008E-06 | 2.898E-04 |
|---|
| positive regulation of phosphorus metabolic process | GO:0010562 |  | 1.11E-06 | 2.836E-04 |
|---|
| response to steroid hormone stimulus | GO:0048545 |  | 1.22E-06 | 2.805E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.339E-06 | 2.799E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to hormone stimulus | GO:0009725 |  | 5.253E-08 | 9.681E-05 |
|---|
| response to corticosteroid stimulus | GO:0031960 |  | 2.132E-06 | 1.965E-03 |
|---|
| endocrine pancreas development | GO:0031018 |  | 4.271E-06 | 2.624E-03 |
|---|
| response to steroid hormone stimulus | GO:0048545 |  | 6.097E-06 | 2.809E-03 |
|---|
| focal adhesion formation | GO:0048041 |  | 8.495E-06 | 3.131E-03 |
|---|
| positive regulation of tyrosine phosphorylation of Stat3 protein | GO:0042517 |  | 1.479E-05 | 4.542E-03 |
|---|
| oxaloacetate metabolic process | GO:0006107 |  | 1.479E-05 | 3.893E-03 |
|---|
| membrane protein ectodomain proteolysis | GO:0006509 |  | 1.479E-05 | 3.406E-03 |
|---|
| pancreas development | GO:0031016 |  | 2.353E-05 | 4.818E-03 |
|---|
| branched chain family amino acid metabolic process | GO:0009081 |  | 2.353E-05 | 4.336E-03 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 2.415E-05 | 4.046E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of locomotion | GO:0040017 |  | 3.1E-07 | 7.13E-04 |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 3.1E-07 | 3.565E-04 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 4.044E-07 | 3.1E-04 |
|---|
| positive regulation of response to external stimulus | GO:0032103 |  | 4.914E-07 | 2.826E-04 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 4.918E-07 | 2.262E-04 |
|---|
| regulation of behavior | GO:0050795 |  | 5.202E-07 | 1.994E-04 |
|---|
| positive regulation of phosphorylation | GO:0042327 |  | 9.143E-07 | 3.004E-04 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.008E-06 | 2.898E-04 |
|---|
| positive regulation of phosphorus metabolic process | GO:0010562 |  | 1.11E-06 | 2.836E-04 |
|---|
| response to steroid hormone stimulus | GO:0048545 |  | 1.22E-06 | 2.805E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.339E-06 | 2.799E-04 |
|---|



