Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6202
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7599 | 3.150e-03 | 4.970e-04 | 7.542e-01 | 1.181e-06 |
|---|
| Loi | 0.2277 | 8.445e-02 | 1.249e-02 | 4.383e-01 | 4.622e-04 |
|---|
| Schmidt | 0.6721 | 0.000e+00 | 0.000e+00 | 4.632e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7904 | 0.000e+00 | 0.000e+00 | 3.514e-03 | 0.000e+00 |
|---|
| Wang | 0.2592 | 3.018e-03 | 4.683e-02 | 4.101e-01 | 5.796e-05 |
|---|
Expression data for subnetwork 6202 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
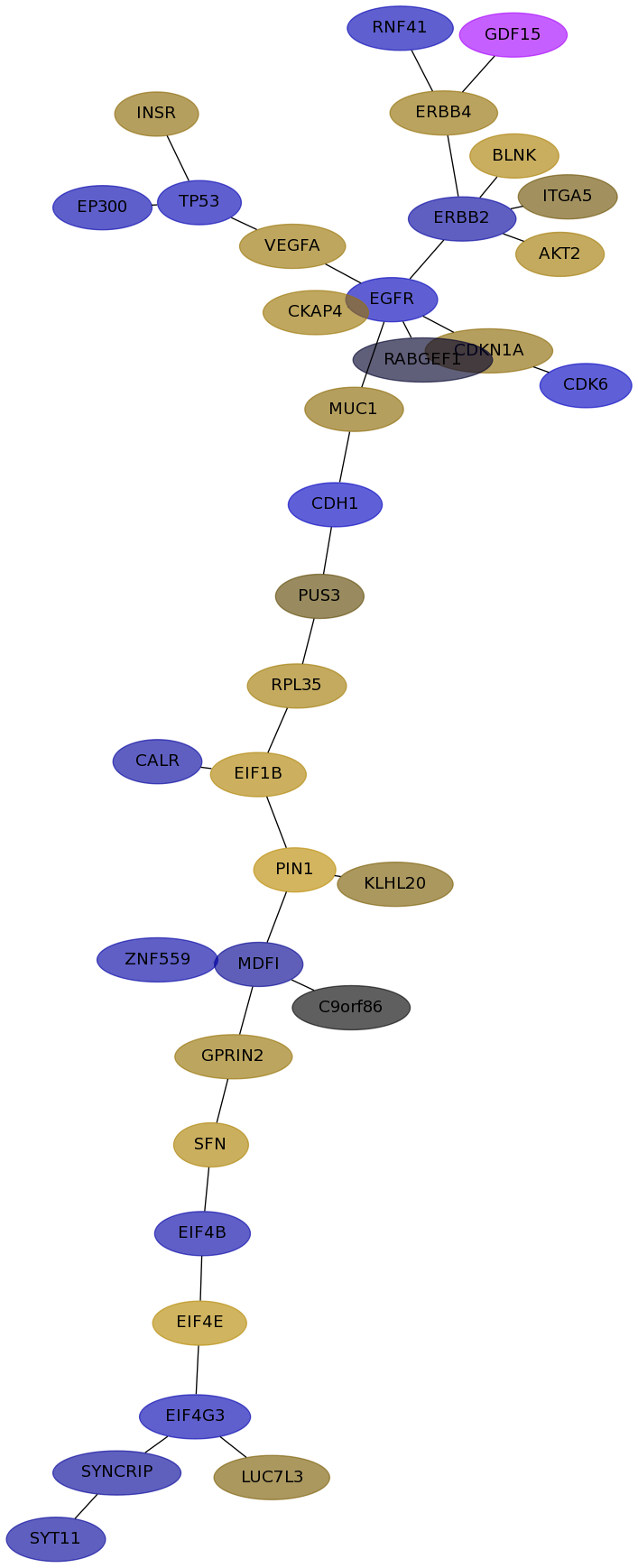
Score for each gene in subnetwork 6202 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| EIF4G3 |   | 1 | 1195 | 2245 | 2246 | -0.158 | 0.203 | -0.089 | -0.028 | 0.039 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| GDF15 |   | 2 | 743 | 658 | 674 | undef | -0.078 | 0.000 | 0.196 | -0.092 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| EIF1B |   | 8 | 222 | 1576 | 1529 | 0.149 | 0.075 | 0.332 | -0.017 | -0.070 |
|---|
| ERBB4 |   | 3 | 557 | 1294 | 1276 | 0.078 | 0.008 | 0.029 | -0.063 | -0.053 |
|---|
| CALR |   | 4 | 440 | 525 | 520 | -0.105 | 0.002 | -0.061 | -0.094 | -0.075 |
|---|
| PUS3 |   | 1 | 1195 | 2245 | 2246 | 0.022 | 0.054 | 0.085 | undef | 0.134 |
|---|
| KLHL20 |   | 2 | 743 | 1274 | 1260 | 0.045 | 0.201 | -0.058 | 0.182 | 0.240 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| GPRIN2 |   | 16 | 104 | 179 | 190 | 0.084 | 0.078 | 0.012 | 0.054 | 0.114 |
|---|
| PIN1 |   | 24 | 62 | 179 | 181 | 0.194 | 0.040 | -0.040 | 0.065 | -0.048 |
|---|
| RNF41 |   | 1 | 1195 | 2245 | 2246 | -0.166 | 0.006 | 0.142 | 0.261 | 0.044 |
|---|
| SYT11 |   | 1 | 1195 | 2245 | 2246 | -0.092 | -0.006 | 0.258 | undef | -0.036 |
|---|
| EIF4E |   | 1 | 1195 | 2245 | 2246 | 0.174 | 0.006 | 0.235 | -0.038 | 0.029 |
|---|
| CDK6 |   | 16 | 104 | 318 | 314 | -0.229 | 0.107 | -0.028 | -0.118 | 0.104 |
|---|
| CKAP4 |   | 6 | 301 | 780 | 764 | 0.093 | 0.152 | -0.167 | 0.315 | -0.017 |
|---|
| MDFI |   | 17 | 95 | 236 | 226 | -0.075 | -0.001 | 0.177 | 0.169 | -0.112 |
|---|
| ZNF559 |   | 3 | 557 | 913 | 908 | -0.122 | 0.089 | 0.145 | -0.005 | -0.014 |
|---|
| RPL35 |   | 8 | 222 | 1323 | 1280 | 0.108 | -0.012 | 0.370 | -0.145 | -0.220 |
|---|
| EIF4B |   | 1 | 1195 | 2245 | 2246 | -0.125 | 0.090 | 0.163 | -0.044 | -0.015 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| ITGA5 |   | 26 | 52 | 179 | 177 | 0.031 | 0.192 | 0.125 | 0.133 | 0.202 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| BLNK |   | 1 | 1195 | 2245 | 2246 | 0.133 | -0.003 | -0.144 | 0.054 | 0.013 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MUC1 |   | 13 | 124 | 1547 | 1490 | 0.063 | 0.214 | -0.190 | undef | -0.132 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| SYNCRIP |   | 2 | 743 | 1364 | 1363 | -0.087 | -0.018 | 0.037 | 0.037 | 0.119 |
|---|
| LUC7L3 |   | 16 | 104 | 296 | 281 | 0.043 | 0.237 | 0.175 | 0.135 | 0.074 |
|---|
| C9orf86 |   | 2 | 743 | 1010 | 1007 | -0.000 | 0.023 | -0.199 | undef | 0.097 |
|---|
| RABGEF1 |   | 2 | 743 | 2197 | 2176 | -0.006 | 0.198 | 0.137 | 0.089 | 0.218 |
|---|
| SFN |   | 25 | 59 | 478 | 454 | 0.141 | 0.270 | 0.023 | -0.102 | 0.101 |
|---|
GO Enrichment output for subnetwork 6202 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of translational initiation | GO:0006446 |  | 2.209E-10 | 5.397E-07 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 1.29E-06 | 1.576E-03 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 7.865E-06 | 6.405E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 8.405E-06 | 5.133E-03 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 9.042E-06 | 4.418E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 9.042E-06 | 3.682E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.868E-05 | 6.52E-03 |
|---|
| response to UV | GO:0009411 |  | 2.024E-05 | 6.182E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 2.175E-05 | 5.904E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.334E-05 | 5.702E-03 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 2.501E-05 | 5.555E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of translational initiation | GO:0006446 |  | 3.94E-10 | 9.479E-07 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 1.676E-06 | 2.016E-03 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.133E-05 | 9.087E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.149E-05 | 6.911E-03 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.297E-05 | 6.242E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.361E-05 | 5.458E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 2.131E-05 | 7.326E-03 |
|---|
| response to UV | GO:0009411 |  | 2.623E-05 | 7.889E-03 |
|---|
| aging | GO:0007568 |  | 3.04E-05 | 8.127E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 3.266E-05 | 7.857E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.504E-05 | 7.663E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of translation | GO:0006417 |  | 1.477E-10 | 3.396E-07 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.826E-08 | 2.1E-05 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 1.368E-06 | 1.049E-03 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.465E-05 | 8.423E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.623E-05 | 7.466E-03 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.839E-05 | 7.049E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.9E-05 | 6.244E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 2.608E-05 | 7.498E-03 |
|---|
| response to UV | GO:0009411 |  | 2.848E-05 | 7.279E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 3.377E-05 | 7.767E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 3.521E-05 | 7.362E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| posttranscriptional regulation of gene expression | GO:0010608 |  | 6.743E-08 | 1.243E-04 |
|---|
| regulation of translation | GO:0006417 |  | 2.046E-07 | 1.885E-04 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.067E-06 | 6.555E-04 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.98E-06 | 1.373E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 3.612E-06 | 1.332E-03 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 4.179E-06 | 1.284E-03 |
|---|
| regulation of MAP kinase activity | GO:0043405 |  | 4.778E-06 | 1.258E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 5.507E-06 | 1.269E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 5.507E-06 | 1.128E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 6.352E-06 | 1.171E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 8.711E-06 | 1.46E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of translation | GO:0006417 |  | 1.477E-10 | 3.396E-07 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.826E-08 | 2.1E-05 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 1.368E-06 | 1.049E-03 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.465E-05 | 8.423E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.623E-05 | 7.466E-03 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.839E-05 | 7.049E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.9E-05 | 6.244E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 2.608E-05 | 7.498E-03 |
|---|
| response to UV | GO:0009411 |  | 2.848E-05 | 7.279E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 3.377E-05 | 7.767E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 3.521E-05 | 7.362E-03 |
|---|



