Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 333-4-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2058 | 1.654e-02 | 1.325e-02 | 1.293e-01 |
|---|
| IPC-NIBC-129 | 0.3253 | 1.222e-02 | 1.849e-02 | 1.557e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2253 | 7.121e-03 | 1.220e-02 | 2.252e-01 |
|---|
| Loi_GPL570 | 0.2173 | 9.254e-02 | 1.035e-01 | 3.491e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1729 | 0.000e+00 | 0.000e+00 | 0.000e+00 |
|---|
| Parker_GPL1390 | 0.2554 | 7.500e-02 | 9.136e-02 | 4.875e-01 |
|---|
| Parker_GPL887 | 0.0706 | 3.471e-01 | 3.658e-01 | 3.766e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3588 | 6.900e-01 | 6.317e-01 | 9.942e-01 |
|---|
| Schmidt | 0.2145 | 2.839e-02 | 3.359e-02 | 2.947e-01 |
|---|
| Sotiriou | 0.2908 | 1.730e-02 | 1.345e-02 | 2.558e-01 |
|---|
| Van-De-Vijver | 0.3584 | 1.888e-03 | 2.715e-03 | 8.914e-02 |
|---|
| Zhang | 0.2493 | 1.104e-01 | 6.888e-02 | 2.387e-01 |
|---|
| Zhou | 0.3847 | 6.719e-02 | 5.564e-02 | 3.947e-01 |
|---|
Expression data for subnetwork 333-4-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
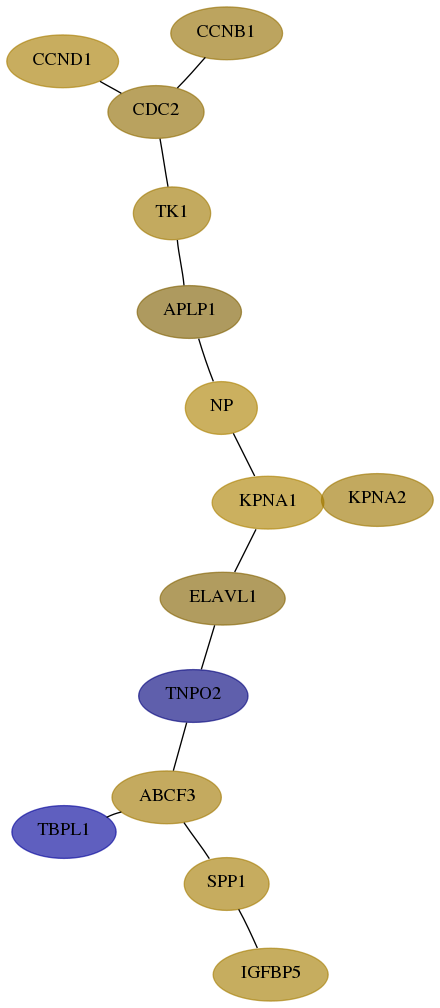
Score for each gene in subnetwork 333-4-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| abcf3 |   | 1 | 116 | 155 | 159 | 0.110 | 0.175 | 0.054 | 0.113 | 0.088 | 0.066 | 0.043 | 0.016 | 0.017 | 0.103 | 0.132 | 0.174 | 0.121 |
|---|
| aplp1 |   | 1 | 116 | 155 | 159 | 0.048 | 0.127 | 0.004 | 0.104 | 0.131 | -0.108 | -0.022 | 0.023 | 0.080 | 0.156 | 0.060 | -0.009 | 0.006 |
|---|
| elavl1 |   | 1 | 116 | 155 | 159 | 0.055 | 0.193 | 0.126 | -0.019 | 0.084 | 0.146 | 0.144 | 0.182 | 0.172 | 0.014 | 0.185 | 0.030 | 0.307 |
|---|
| spp1 |   | 3 | 62 | 106 | 95 | 0.121 | 0.316 | 0.018 | -0.074 | 0.132 | 0.124 | -0.237 | 0.185 | 0.031 | 0.109 | 0.212 | 0.141 | 0.275 |
|---|
| np |   | 1 | 116 | 155 | 159 | 0.143 | 0.122 | 0.152 | 0.103 | 0.104 | 0.205 | 0.440 | 0.234 | 0.143 | 0.098 | 0.188 | -0.029 | 0.209 |
|---|
| ccnb1 |   | 42 | 8 | 56 | 39 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | undef | 0.167 | 0.398 |
|---|
| kpna1 |   | 2 | 78 | 153 | 146 | 0.147 | 0.079 | 0.132 | -0.037 | 0.109 | 0.198 | -0.559 | 0.256 | 0.098 | 0.096 | 0.109 | 0.147 | 0.107 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| tnpo2 |   | 1 | 116 | 155 | 159 | -0.046 | 0.070 | -0.065 | 0.117 | 0.071 | 0.186 | 0.471 | 0.056 | 0.166 | 0.038 | -0.020 | 0.085 | -0.110 |
|---|
| tbpl1 |   | 1 | 116 | 155 | 159 | -0.093 | 0.349 | 0.069 | 0.042 | 0.056 | 0.239 | -0.016 | 0.021 | 0.098 | -0.123 | -0.010 | 0.128 | 0.207 |
|---|
| igfbp5 |   | 2 | 78 | 112 | 109 | 0.123 | 0.143 | 0.025 | 0.150 | 0.143 | 0.006 | -0.178 | -0.055 | 0.063 | 0.147 | 0.238 | 0.026 | -0.043 |
|---|
| tk1 |   | 7 | 30 | 84 | 67 | 0.109 | 0.149 | 0.190 | -0.017 | 0.131 | 0.105 | -0.100 | 0.257 | 0.204 | 0.142 | 0.293 | 0.164 | 0.222 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| kpna2 |   | 1 | 116 | 155 | 159 | 0.105 | 0.283 | 0.149 | 0.077 | 0.163 | 0.238 | 0.085 | 0.314 | 0.228 | 0.151 | 0.220 | 0.170 | 0.305 |
|---|
GO Enrichment output for subnetwork 333-4-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| NLS-bearing substrate import into nucleus | GO:0006607 |  | 2.515E-06 | 5.786E-03 |
|---|
| regulation of DNA recombination | GO:0000018 |  | 5.94E-06 | 6.831E-03 |
|---|
| protein import into nucleus | GO:0006606 |  | 7.457E-06 | 5.717E-03 |
|---|
| nuclear import | GO:0051170 |  | 7.868E-06 | 4.524E-03 |
|---|
| protein localization in nucleus | GO:0034504 |  | 1.018E-05 | 4.681E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.357E-05 | 5.201E-03 |
|---|
| interphase | GO:0051325 |  | 1.486E-05 | 4.883E-03 |
|---|
| protein import | GO:0017038 |  | 2.368E-05 | 6.808E-03 |
|---|
| protein localization in organelle | GO:0033365 |  | 2.464E-05 | 6.296E-03 |
|---|
| mRNA stabilization | GO:0048255 |  | 4.252E-05 | 9.78E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.252E-05 | 8.891E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| NLS-bearing substrate import into nucleus | GO:0006607 |  | 5.295E-07 | 1.293E-03 |
|---|
| protein import into nucleus | GO:0006606 |  | 1.465E-06 | 1.789E-03 |
|---|
| nuclear import | GO:0051170 |  | 1.538E-06 | 1.252E-03 |
|---|
| regulation of DNA recombination | GO:0000018 |  | 1.666E-06 | 1.018E-03 |
|---|
| protein localization in nucleus | GO:0034504 |  | 2.035E-06 | 9.941E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.991E-06 | 1.218E-03 |
|---|
| deoxyribonucleoside triphosphate metabolic process | GO:0009200 |  | 9.954E-06 | 3.474E-03 |
|---|
| deoxyribonucleoside metabolic process | GO:0009120 |  | 1.492E-05 | 4.557E-03 |
|---|
| nucleoside catabolic process | GO:0009164 |  | 1.492E-05 | 4.051E-03 |
|---|
| G2 phase | GO:0051319 |  | 1.492E-05 | 3.645E-03 |
|---|
| mRNA stabilization | GO:0048255 |  | 2.088E-05 | 4.637E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| NLS-bearing substrate import into nucleus | GO:0006607 |  | 7.288E-07 | 1.753E-03 |
|---|
| protein import into nucleus | GO:0006606 |  | 2.014E-06 | 2.423E-03 |
|---|
| nuclear import | GO:0051170 |  | 2.117E-06 | 1.698E-03 |
|---|
| regulation of DNA recombination | GO:0000018 |  | 2.292E-06 | 1.379E-03 |
|---|
| protein localization in nucleus | GO:0034504 |  | 2.819E-06 | 1.357E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.006E-06 | 1.607E-03 |
|---|
| interphase | GO:0051325 |  | 4.535E-06 | 1.559E-03 |
|---|
| deoxyribonucleoside triphosphate metabolic process | GO:0009200 |  | 1.234E-05 | 3.712E-03 |
|---|
| deoxyribonucleoside metabolic process | GO:0009120 |  | 1.85E-05 | 4.946E-03 |
|---|
| nucleoside catabolic process | GO:0009164 |  | 1.85E-05 | 4.452E-03 |
|---|
| G2 phase | GO:0051319 |  | 1.85E-05 | 4.047E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| NLS-bearing substrate import into nucleus | GO:0006607 |  | 5.295E-07 | 1.293E-03 |
|---|
| protein import into nucleus | GO:0006606 |  | 1.465E-06 | 1.789E-03 |
|---|
| nuclear import | GO:0051170 |  | 1.538E-06 | 1.252E-03 |
|---|
| regulation of DNA recombination | GO:0000018 |  | 1.666E-06 | 1.018E-03 |
|---|
| protein localization in nucleus | GO:0034504 |  | 2.035E-06 | 9.941E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.991E-06 | 1.218E-03 |
|---|
| deoxyribonucleoside triphosphate metabolic process | GO:0009200 |  | 9.954E-06 | 3.474E-03 |
|---|
| deoxyribonucleoside metabolic process | GO:0009120 |  | 1.492E-05 | 4.557E-03 |
|---|
| nucleoside catabolic process | GO:0009164 |  | 1.492E-05 | 4.051E-03 |
|---|
| G2 phase | GO:0051319 |  | 1.492E-05 | 3.645E-03 |
|---|
| mRNA stabilization | GO:0048255 |  | 2.088E-05 | 4.637E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| NLS-bearing substrate import into nucleus | GO:0006607 |  | 7.288E-07 | 1.753E-03 |
|---|
| protein import into nucleus | GO:0006606 |  | 2.014E-06 | 2.423E-03 |
|---|
| nuclear import | GO:0051170 |  | 2.117E-06 | 1.698E-03 |
|---|
| regulation of DNA recombination | GO:0000018 |  | 2.292E-06 | 1.379E-03 |
|---|
| protein localization in nucleus | GO:0034504 |  | 2.819E-06 | 1.357E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.006E-06 | 1.607E-03 |
|---|
| interphase | GO:0051325 |  | 4.535E-06 | 1.559E-03 |
|---|
| deoxyribonucleoside triphosphate metabolic process | GO:0009200 |  | 1.234E-05 | 3.712E-03 |
|---|
| deoxyribonucleoside metabolic process | GO:0009120 |  | 1.85E-05 | 4.946E-03 |
|---|
| nucleoside catabolic process | GO:0009164 |  | 1.85E-05 | 4.452E-03 |
|---|
| G2 phase | GO:0051319 |  | 1.85E-05 | 4.047E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear localization sequence binding | GO:0008139 |  | 8.244E-09 | 2.944E-05 |
|---|
| signal sequence binding | GO:0005048 |  | 2.789E-07 | 4.979E-04 |
|---|
| mRNA stabilization | GO:0048255 |  | 6.073E-06 | 7.228E-03 |
|---|
| insulin-like growth factor I binding | GO:0031994 |  | 6.073E-06 | 5.421E-03 |
|---|
| deoxyribonucleoside metabolic process | GO:0009120 |  | 6.073E-06 | 4.337E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 9.105E-06 | 5.419E-03 |
|---|
| nucleoside catabolic process | GO:0009164 |  | 9.105E-06 | 4.645E-03 |
|---|
| nucleoside metabolic process | GO:0009116 |  | 9.45E-06 | 4.218E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.274E-05 | 5.055E-03 |
|---|
| nucleobase. nucleoside and nucleotide catabolic process | GO:0034656 |  | 1.274E-05 | 4.55E-03 |
|---|
| negative regulation of smooth muscle cell proliferation | GO:0048662 |  | 1.274E-05 | 4.136E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear localization sequence binding | GO:0008139 |  | 1.02E-10 | 3.721E-07 |
|---|
| signal sequence binding | GO:0005048 |  | 6.871E-09 | 1.253E-05 |
|---|
| NLS-bearing substrate import into nucleus | GO:0006607 |  | 6.74E-07 | 8.195E-04 |
|---|
| nuclear pore | GO:0005643 |  | 1.544E-06 | 1.408E-03 |
|---|
| protein import into nucleus | GO:0006606 |  | 1.987E-06 | 1.45E-03 |
|---|
| nuclear import | GO:0051170 |  | 2.111E-06 | 1.284E-03 |
|---|
| regulation of DNA recombination | GO:0000018 |  | 2.271E-06 | 1.183E-03 |
|---|
| protein localization in nucleus | GO:0034504 |  | 2.666E-06 | 1.216E-03 |
|---|
| pore complex | GO:0046930 |  | 2.82E-06 | 1.143E-03 |
|---|
| protein transporter activity | GO:0008565 |  | 2.981E-06 | 1.087E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.523E-06 | 1.5E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| NLS-bearing substrate import into nucleus | GO:0006607 |  | 7.288E-07 | 1.753E-03 |
|---|
| protein import into nucleus | GO:0006606 |  | 2.014E-06 | 2.423E-03 |
|---|
| nuclear import | GO:0051170 |  | 2.117E-06 | 1.698E-03 |
|---|
| regulation of DNA recombination | GO:0000018 |  | 2.292E-06 | 1.379E-03 |
|---|
| protein localization in nucleus | GO:0034504 |  | 2.819E-06 | 1.357E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.006E-06 | 1.607E-03 |
|---|
| interphase | GO:0051325 |  | 4.535E-06 | 1.559E-03 |
|---|
| deoxyribonucleoside triphosphate metabolic process | GO:0009200 |  | 1.234E-05 | 3.712E-03 |
|---|
| deoxyribonucleoside metabolic process | GO:0009120 |  | 1.85E-05 | 4.946E-03 |
|---|
| nucleoside catabolic process | GO:0009164 |  | 1.85E-05 | 4.452E-03 |
|---|
| G2 phase | GO:0051319 |  | 1.85E-05 | 4.047E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| NLS-bearing substrate import into nucleus | GO:0006607 |  | 2.515E-06 | 5.786E-03 |
|---|
| regulation of DNA recombination | GO:0000018 |  | 5.94E-06 | 6.831E-03 |
|---|
| protein import into nucleus | GO:0006606 |  | 7.457E-06 | 5.717E-03 |
|---|
| nuclear import | GO:0051170 |  | 7.868E-06 | 4.524E-03 |
|---|
| protein localization in nucleus | GO:0034504 |  | 1.018E-05 | 4.681E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.357E-05 | 5.201E-03 |
|---|
| interphase | GO:0051325 |  | 1.486E-05 | 4.883E-03 |
|---|
| protein import | GO:0017038 |  | 2.368E-05 | 6.808E-03 |
|---|
| protein localization in organelle | GO:0033365 |  | 2.464E-05 | 6.296E-03 |
|---|
| mRNA stabilization | GO:0048255 |  | 4.252E-05 | 9.78E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.252E-05 | 8.891E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| NLS-bearing substrate import into nucleus | GO:0006607 |  | 2.515E-06 | 5.786E-03 |
|---|
| regulation of DNA recombination | GO:0000018 |  | 5.94E-06 | 6.831E-03 |
|---|
| protein import into nucleus | GO:0006606 |  | 7.457E-06 | 5.717E-03 |
|---|
| nuclear import | GO:0051170 |  | 7.868E-06 | 4.524E-03 |
|---|
| protein localization in nucleus | GO:0034504 |  | 1.018E-05 | 4.681E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.357E-05 | 5.201E-03 |
|---|
| interphase | GO:0051325 |  | 1.486E-05 | 4.883E-03 |
|---|
| protein import | GO:0017038 |  | 2.368E-05 | 6.808E-03 |
|---|
| protein localization in organelle | GO:0033365 |  | 2.464E-05 | 6.296E-03 |
|---|
| mRNA stabilization | GO:0048255 |  | 4.252E-05 | 9.78E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.252E-05 | 8.891E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| NLS-bearing substrate import into nucleus | GO:0006607 |  | 1.061E-07 | 1.955E-04 |
|---|
| regulation of DNA recombination | GO:0000018 |  | 8.623E-07 | 7.946E-04 |
|---|
| posttranscriptional regulation of gene expression | GO:0010608 |  | 1.183E-05 | 7.27E-03 |
|---|
| nucleoside metabolic process | GO:0009116 |  | 1.334E-05 | 6.147E-03 |
|---|
| mRNA stabilization | GO:0048255 |  | 2.318E-05 | 8.545E-03 |
|---|
| nucleobase. nucleoside and nucleotide catabolic process | GO:0034656 |  | 2.318E-05 | 7.121E-03 |
|---|
| pyrimidine nucleoside triphosphate biosynthetic process | GO:0009148 |  | 3.474E-05 | 9.147E-03 |
|---|
| protein import into nucleus | GO:0006606 |  | 3.886E-05 | 8.953E-03 |
|---|
| nuclear import | GO:0051170 |  | 4.16E-05 | 8.519E-03 |
|---|
| regulation of mRNA stability | GO:0043488 |  | 4.86E-05 | 8.956E-03 |
|---|
| protein localization in nucleus | GO:0034504 |  | 5.057E-05 | 8.473E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| NLS-bearing substrate import into nucleus | GO:0006607 |  | 2.515E-06 | 5.786E-03 |
|---|
| regulation of DNA recombination | GO:0000018 |  | 5.94E-06 | 6.831E-03 |
|---|
| protein import into nucleus | GO:0006606 |  | 7.457E-06 | 5.717E-03 |
|---|
| nuclear import | GO:0051170 |  | 7.868E-06 | 4.524E-03 |
|---|
| protein localization in nucleus | GO:0034504 |  | 1.018E-05 | 4.681E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.357E-05 | 5.201E-03 |
|---|
| interphase | GO:0051325 |  | 1.486E-05 | 4.883E-03 |
|---|
| protein import | GO:0017038 |  | 2.368E-05 | 6.808E-03 |
|---|
| protein localization in organelle | GO:0033365 |  | 2.464E-05 | 6.296E-03 |
|---|
| mRNA stabilization | GO:0048255 |  | 4.252E-05 | 9.78E-03 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 4.252E-05 | 8.891E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| NLS-bearing substrate import into nucleus | GO:0006607 |  | 2.754E-06 | 6.536E-03 |
|---|
| regulation of DNA recombination | GO:0000018 |  | 5.176E-06 | 6.141E-03 |
|---|
| protein import into nucleus | GO:0006606 |  | 8.953E-06 | 7.082E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 9.821E-06 | 5.826E-03 |
|---|
| interphase | GO:0051325 |  | 1.075E-05 | 5.101E-03 |
|---|
| nuclear import | GO:0051170 |  | 1.124E-05 | 4.444E-03 |
|---|
| protein localization in nucleus | GO:0034504 |  | 1.226E-05 | 4.156E-03 |
|---|
| deoxyribonucleoside metabolic process | GO:0009120 |  | 3.513E-05 | 0.01042154 |
|---|
| nucleoside catabolic process | GO:0009164 |  | 3.513E-05 | 9.264E-03 |
|---|
| G2 phase | GO:0051319 |  | 3.513E-05 | 8.337E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.914E-05 | 0.01060071 |
|---|



