Study run-a2
Study informations
103 subnetworks in total page | file
306 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 2932-3-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2032 | 1.554e-02 | 1.235e-02 | 1.232e-01 |
|---|
| IPC-NIBC-129 | 0.2687 | 3.426e-02 | 5.010e-02 | 3.464e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2284 | 5.904e-03 | 1.028e-02 | 2.020e-01 |
|---|
| Loi_GPL570 | 0.2520 | 5.695e-02 | 6.457e-02 | 2.421e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1661 | 1.000e-06 | 9.000e-06 | 1.000e-06 |
|---|
| Parker_GPL1390 | 0.2640 | 6.401e-02 | 7.859e-02 | 4.495e-01 |
|---|
| Parker_GPL887 | 0.1055 | 3.526e-01 | 3.713e-01 | 3.834e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3434 | 5.157e-01 | 4.434e-01 | 9.704e-01 |
|---|
| Schmidt | 0.2257 | 1.030e-02 | 1.136e-02 | 1.464e-01 |
|---|
| Sotiriou | 0.3004 | 1.475e-02 | 1.142e-02 | 2.345e-01 |
|---|
| Van-De-Vijver | 0.2844 | 1.226e-03 | 1.765e-03 | 6.724e-02 |
|---|
| Zhang | 0.3108 | 1.224e-01 | 7.842e-02 | 2.601e-01 |
|---|
| Zhou | 0.4089 | 7.731e-02 | 6.673e-02 | 4.366e-01 |
|---|
Expression data for subnetwork 2932-3-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Van-De-Vijver |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
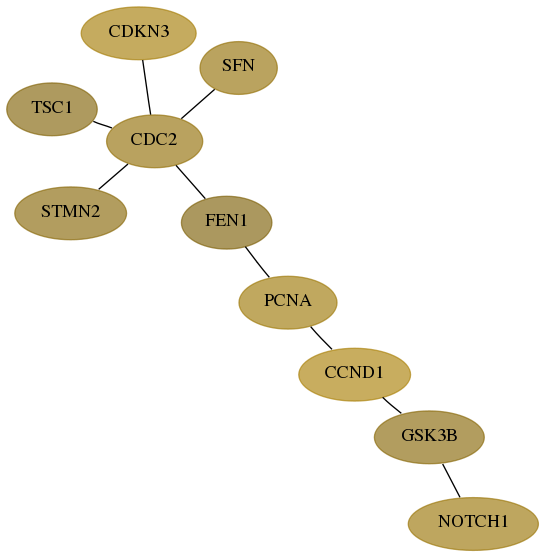
Score for each gene in subnetwork 2932-3-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Van-De-Vijver | Zhang | Zhou |
|---|
| cdkn3 |   | 53 | 5 | 29 | 22 | 0.114 | 0.269 | 0.199 | 0.136 | 0.193 | 0.166 | -0.009 | 0.323 | 0.186 | 0.198 | 0.201 | 0.271 | 0.372 |
|---|
| pcna |   | 15 | 11 | 110 | 85 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.119 | 0.104 | 0.256 | 0.151 | 0.059 | 0.201 | 0.123 | 0.233 |
|---|
| fen1 |   | 9 | 22 | 133 | 112 | 0.045 | 0.194 | 0.169 | 0.196 | 0.136 | 0.252 | 0.033 | 0.332 | 0.221 | 0.144 | 0.242 | 0.056 | 0.114 |
|---|
| stmn2 |   | 29 | 9 | 34 | 27 | 0.057 | 0.106 | 0.005 | 0.052 | 0.110 | 0.071 | -0.099 | 0.006 | 0.034 | 0.103 | 0.008 | 0.113 | -0.039 |
|---|
| notch1 |   | 3 | 62 | 133 | 125 | 0.088 | 0.293 | -0.017 | -0.179 | 0.066 | 0.172 | 0.176 | 0.061 | 0.087 | 0.030 | 0.082 | -0.062 | 0.218 |
|---|
| gsk3b |   | 2 | 78 | 133 | 129 | 0.057 | 0.003 | 0.058 | 0.029 | 0.154 | 0.025 | -0.462 | -0.068 | 0.119 | 0.070 | 0.188 | 0.146 | 0.093 |
|---|
| ccnd1 |   | 92 | 2 | 18 | 12 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | undef | 0.144 | 0.290 |
|---|
| tsc1 |   | 58 | 4 | 18 | 13 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.095 | -0.007 | 0.207 |
|---|
| cdc2 |   | 96 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.162 | 0.297 | 0.336 |
|---|
| sfn |   | 61 | 3 | 1 | 2 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.158 | 0.079 | 0.181 |
|---|
GO Enrichment output for subnetwork 2932-3-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.293E-08 | 1.447E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 5.392E-07 | 6.2E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 7.173E-07 | 5.499E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.951E-06 | 2.847E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.947E-06 | 2.736E-03 |
|---|
| anagen | GO:0042640 |  | 5.947E-06 | 2.28E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 5.947E-06 | 1.954E-03 |
|---|
| ectodermal gut morphogenesis | GO:0048567 |  | 5.947E-06 | 1.71E-03 |
|---|
| response to UV | GO:0009411 |  | 6.081E-06 | 1.554E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 8.917E-06 | 2.051E-03 |
|---|
| ER overload response | GO:0006983 |  | 8.917E-06 | 1.864E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.066E-08 | 2.604E-05 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.933E-07 | 2.362E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 2.715E-07 | 2.211E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.123E-06 | 6.856E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.898E-06 | 9.275E-04 |
|---|
| anagen | GO:0042640 |  | 1.898E-06 | 7.729E-04 |
|---|
| regulation of keratinocyte differentiation | GO:0045616 |  | 1.898E-06 | 6.625E-04 |
|---|
| stem cell division | GO:0017145 |  | 1.898E-06 | 5.797E-04 |
|---|
| response to UV | GO:0009411 |  | 1.935E-06 | 5.253E-04 |
|---|
| hair follicle maturation | GO:0048820 |  | 2.847E-06 | 6.954E-04 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 2.847E-06 | 6.322E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.549E-08 | 3.727E-05 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.284E-07 | 2.747E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 3.25E-07 | 2.606E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.498E-06 | 9.009E-04 |
|---|
| response to UV | GO:0009411 |  | 2.307E-06 | 1.11E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.313E-06 | 9.276E-04 |
|---|
| anagen | GO:0042640 |  | 2.313E-06 | 7.951E-04 |
|---|
| regulation of keratinocyte differentiation | GO:0045616 |  | 2.313E-06 | 6.957E-04 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 2.313E-06 | 6.184E-04 |
|---|
| stem cell division | GO:0017145 |  | 2.313E-06 | 5.566E-04 |
|---|
| hair follicle maturation | GO:0048820 |  | 3.469E-06 | 7.588E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.066E-08 | 2.604E-05 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.933E-07 | 2.362E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 2.715E-07 | 2.211E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.123E-06 | 6.856E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.898E-06 | 9.275E-04 |
|---|
| anagen | GO:0042640 |  | 1.898E-06 | 7.729E-04 |
|---|
| regulation of keratinocyte differentiation | GO:0045616 |  | 1.898E-06 | 6.625E-04 |
|---|
| stem cell division | GO:0017145 |  | 1.898E-06 | 5.797E-04 |
|---|
| response to UV | GO:0009411 |  | 1.935E-06 | 5.253E-04 |
|---|
| hair follicle maturation | GO:0048820 |  | 2.847E-06 | 6.954E-04 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 2.847E-06 | 6.322E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.549E-08 | 3.727E-05 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.284E-07 | 2.747E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 3.25E-07 | 2.606E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.498E-06 | 9.009E-04 |
|---|
| response to UV | GO:0009411 |  | 2.307E-06 | 1.11E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.313E-06 | 9.276E-04 |
|---|
| anagen | GO:0042640 |  | 2.313E-06 | 7.951E-04 |
|---|
| regulation of keratinocyte differentiation | GO:0045616 |  | 2.313E-06 | 6.957E-04 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 2.313E-06 | 6.184E-04 |
|---|
| stem cell division | GO:0017145 |  | 2.313E-06 | 5.566E-04 |
|---|
| hair follicle maturation | GO:0048820 |  | 3.469E-06 | 7.588E-04 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 8.812E-09 | 3.147E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.46E-08 | 2.607E-05 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 4.069E-07 | 4.843E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 5.124E-07 | 4.574E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.361E-06 | 1.686E-03 |
|---|
| response to UV | GO:0009411 |  | 2.863E-06 | 1.704E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 3.314E-06 | 1.69E-03 |
|---|
| MutLalpha complex binding | GO:0032405 |  | 3.314E-06 | 1.479E-03 |
|---|
| ribonuclease H activity | GO:0004523 |  | 3.314E-06 | 1.315E-03 |
|---|
| negative regulation of myoblast differentiation | GO:0045662 |  | 3.314E-06 | 1.183E-03 |
|---|
| endodeoxyribonuclease activity. producing 5'-phosphomonoesters | GO:0016888 |  | 3.314E-06 | 1.076E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 8.439E-09 | 3.078E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.59E-08 | 2.899E-05 |
|---|
| growth cone | GO:0030426 |  | 3.034E-07 | 3.69E-04 |
|---|
| site of polarized growth | GO:0030427 |  | 3.447E-07 | 3.144E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 3.895E-07 | 2.842E-04 |
|---|
| fat cell differentiation | GO:0045444 |  | 4.381E-07 | 2.664E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 5.469E-07 | 2.85E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.839E-06 | 8.386E-04 |
|---|
| response to UV | GO:0009411 |  | 2.113E-06 | 8.565E-04 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 3.136E-06 | 1.144E-03 |
|---|
| ribonuclease H activity | GO:0004523 |  | 3.136E-06 | 1.04E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.549E-08 | 3.727E-05 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 2.284E-07 | 2.747E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 3.25E-07 | 2.606E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.498E-06 | 9.009E-04 |
|---|
| response to UV | GO:0009411 |  | 2.307E-06 | 1.11E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.313E-06 | 9.276E-04 |
|---|
| anagen | GO:0042640 |  | 2.313E-06 | 7.951E-04 |
|---|
| regulation of keratinocyte differentiation | GO:0045616 |  | 2.313E-06 | 6.957E-04 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 2.313E-06 | 6.184E-04 |
|---|
| stem cell division | GO:0017145 |  | 2.313E-06 | 5.566E-04 |
|---|
| hair follicle maturation | GO:0048820 |  | 3.469E-06 | 7.588E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.293E-08 | 1.447E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 5.392E-07 | 6.2E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 7.173E-07 | 5.499E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.951E-06 | 2.847E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.947E-06 | 2.736E-03 |
|---|
| anagen | GO:0042640 |  | 5.947E-06 | 2.28E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 5.947E-06 | 1.954E-03 |
|---|
| ectodermal gut morphogenesis | GO:0048567 |  | 5.947E-06 | 1.71E-03 |
|---|
| response to UV | GO:0009411 |  | 6.081E-06 | 1.554E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 8.917E-06 | 2.051E-03 |
|---|
| ER overload response | GO:0006983 |  | 8.917E-06 | 1.864E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.293E-08 | 1.447E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 5.392E-07 | 6.2E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 7.173E-07 | 5.499E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.951E-06 | 2.847E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.947E-06 | 2.736E-03 |
|---|
| anagen | GO:0042640 |  | 5.947E-06 | 2.28E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 5.947E-06 | 1.954E-03 |
|---|
| ectodermal gut morphogenesis | GO:0048567 |  | 5.947E-06 | 1.71E-03 |
|---|
| response to UV | GO:0009411 |  | 6.081E-06 | 1.554E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 8.917E-06 | 2.051E-03 |
|---|
| ER overload response | GO:0006983 |  | 8.917E-06 | 1.864E-03 |
|---|
Van-De-Vijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| hair follicle maturation | GO:0048820 |  | 1.181E-05 | 0.02176916 |
|---|
| regulation of myoblast differentiation | GO:0045661 |  | 1.181E-05 | 0.01088458 |
|---|
| positive regulation of epidermis development | GO:0045684 |  | 1.181E-05 | 7.256E-03 |
|---|
| regulation of epidermal cell differentiation | GO:0045604 |  | 1.181E-05 | 5.442E-03 |
|---|
| ectodermal gut morphogenesis | GO:0048567 |  | 1.181E-05 | 4.354E-03 |
|---|
| phosphoinositide-mediated signaling | GO:0048015 |  | 1.431E-05 | 4.397E-03 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 1.771E-05 | 4.662E-03 |
|---|
| ER overload response | GO:0006983 |  | 1.771E-05 | 4.079E-03 |
|---|
| peptidyl-serine phosphorylation | GO:0018105 |  | 1.771E-05 | 3.626E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.771E-05 | 3.263E-03 |
|---|
| regulation of Rho GTPase activity | GO:0032319 |  | 1.771E-05 | 2.967E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.293E-08 | 1.447E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 5.392E-07 | 6.2E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 7.173E-07 | 5.499E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.951E-06 | 2.847E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.947E-06 | 2.736E-03 |
|---|
| anagen | GO:0042640 |  | 5.947E-06 | 2.28E-03 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 5.947E-06 | 1.954E-03 |
|---|
| ectodermal gut morphogenesis | GO:0048567 |  | 5.947E-06 | 1.71E-03 |
|---|
| response to UV | GO:0009411 |  | 6.081E-06 | 1.554E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 8.917E-06 | 2.051E-03 |
|---|
| ER overload response | GO:0006983 |  | 8.917E-06 | 1.864E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.042E-07 | 2.474E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.038E-06 | 1.232E-03 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.332E-06 | 1.054E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.723E-06 | 3.988E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 6.867E-06 | 3.259E-03 |
|---|
| anagen | GO:0042640 |  | 6.867E-06 | 2.716E-03 |
|---|
| negative regulation of myoblast differentiation | GO:0045662 |  | 6.867E-06 | 2.328E-03 |
|---|
| regulation of keratinocyte differentiation | GO:0045616 |  | 6.867E-06 | 2.037E-03 |
|---|
| ectodermal gut morphogenesis | GO:0048567 |  | 6.867E-06 | 1.811E-03 |
|---|
| response to UV | GO:0009411 |  | 8.218E-06 | 1.95E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.029E-05 | 2.221E-03 |
|---|



