Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 374291-4-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2111 | 7.693e-03 | 9.179e-03 | 8.129e-02 |
|---|
| IPC-NIBC-129 | 0.2299 | 5.053e-02 | 6.437e-02 | 4.555e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2610 | 3.130e-03 | 5.417e-03 | 1.244e-01 |
|---|
| Loi_GPL570 | 0.2970 | 8.072e-02 | 8.021e-02 | 3.584e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1646 | 2.000e-06 | 1.500e-05 | 6.000e-06 |
|---|
| Parker_GPL1390 | 0.2575 | 8.411e-02 | 8.257e-02 | 4.967e-01 |
|---|
| Parker_GPL887 | 0.2356 | 2.834e-01 | 3.168e-01 | 2.847e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3418 | 1.571e-01 | 1.235e-01 | 6.655e-01 |
|---|
| Schmidt | 0.2625 | 3.146e-03 | 4.714e-03 | 5.103e-02 |
|---|
| Sotiriou | 0.3853 | 2.878e-03 | 8.640e-04 | 6.897e-02 |
|---|
| Wang | 0.0710 | 1.029e-01 | 7.108e-02 | 3.723e-01 |
|---|
| Zhang | 0.2267 | 4.689e-01 | 4.144e-01 | 7.752e-01 |
|---|
| Zhou | 0.4044 | 7.291e-02 | 7.837e-02 | 4.368e-01 |
|---|
Expression data for subnetwork 374291-4-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
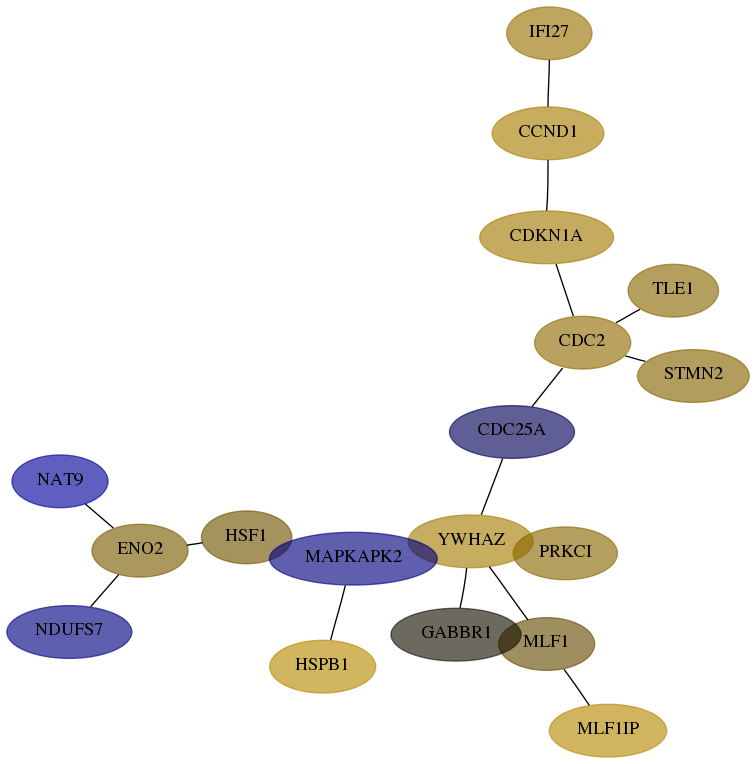
Score for each gene in subnetwork 374291-4-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| stmn2 |   | 29 | 8 | 1 | 3 | 0.057 | 0.106 | 0.005 | 0.052 | 0.110 | 0.071 | -0.099 | 0.006 | 0.034 | 0.103 | 0.075 | 0.113 | -0.039 |
|---|
| ndufs7 |   | 1 | 164 | 109 | 126 | -0.053 | 0.081 | 0.127 | 0.168 | 0.071 | 0.009 | -0.032 | 0.192 | 0.081 | -0.002 | -0.034 | 0.061 | 0.026 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| tle1 |   | 23 | 9 | 78 | 60 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | -0.028 | 0.068 | 0.025 |
|---|
| mlf1ip |   | 5 | 50 | 53 | 47 | 0.182 | 0.147 | 0.186 | 0.144 | 0.242 | 0.266 | 0.005 | 0.345 | 0.182 | 0.197 | 0.172 | 0.240 | 0.372 |
|---|
| prkci |   | 12 | 20 | 31 | 19 | 0.065 | 0.104 | 0.012 | 0.170 | 0.112 | 0.088 | -0.103 | -0.170 | 0.196 | 0.128 | -0.054 | 0.124 | 0.149 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| hsf1 |   | 1 | 164 | 109 | 126 | 0.035 | 0.032 | 0.040 | 0.244 | 0.098 | 0.100 | 0.319 | 0.146 | 0.165 | 0.116 | 0.118 | 0.100 | 0.086 |
|---|
| gabbr1 |   | 6 | 43 | 88 | 77 | 0.003 | -0.016 | -0.022 | -0.051 | 0.082 | 0.119 | 0.584 | -0.082 | 0.049 | 0.057 | 0.011 | 0.096 | 0.116 |
|---|
| mlf1 |   | 5 | 50 | 53 | 47 | 0.027 | 0.224 | 0.053 | 0.047 | 0.169 | 0.128 | 0.021 | -0.028 | 0.106 | 0.176 | 0.126 | 0.098 | 0.141 |
|---|
| nat9 |   | 1 | 164 | 109 | 126 | -0.093 | 0.106 | 0.107 | 0.056 | 0.136 | 0.067 | 0.173 | 0.137 | 0.082 | 0.186 | -0.023 | 0.122 | 0.109 |
|---|
| ywhaz |   | 11 | 21 | 31 | 20 | 0.124 | 0.089 | 0.087 | 0.200 | 0.141 | 0.110 | 0.228 | 0.199 | 0.056 | 0.123 | -0.026 | 0.103 | 0.213 |
|---|
| hspb1 |   | 19 | 11 | 17 | 11 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | -0.026 | -0.065 | 0.289 | -0.021 | 0.201 | 0.125 | -0.077 | 0.290 |
|---|
| eno2 |   | 1 | 164 | 109 | 126 | 0.045 | 0.124 | 0.044 | 0.231 | 0.141 | -0.139 | 0.514 | 0.026 | 0.017 | 0.151 | 0.167 | 0.086 | 0.139 |
|---|
| cdc25a |   | 9 | 27 | 109 | 90 | -0.019 | 0.221 | 0.093 | 0.158 | 0.120 | -0.061 | 0.406 | 0.207 | 0.211 | 0.016 | 0.069 | 0.183 | 0.264 |
|---|
| mapkapk2 |   | 13 | 19 | 17 | 12 | -0.050 | 0.102 | 0.156 | -0.116 | 0.090 | 0.168 | 0.303 | 0.074 | 0.171 | 0.117 | 0.085 | -0.124 | 0.312 |
|---|
| ifi27 |   | 2 | 104 | 109 | 110 | 0.089 | 0.086 | 0.105 | 0.055 | 0.101 | 0.007 | 0.360 | 0.149 | 0.129 | 0.045 | -0.074 | 0.118 | 0.194 |
|---|
GO Enrichment output for subnetwork 374291-4-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 6.703E-08 | 1.542E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.097E-06 | 1.262E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.793E-06 | 2.141E-03 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.699E-06 | 2.127E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 7.491E-06 | 3.446E-03 |
|---|
| sperm motility | GO:0030317 |  | 1.323E-05 | 5.071E-03 |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.698E-05 | 5.581E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 2.372E-05 | 6.821E-03 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 3.08E-05 | 7.871E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 3.208E-05 | 7.377E-03 |
|---|
| hemopoietic progenitor cell differentiation | GO:0002244 |  | 3.658E-05 | 7.649E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.531E-08 | 3.741E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.351E-07 | 5.315E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.355E-07 | 5.175E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 8.499E-07 | 5.191E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 2.01E-06 | 9.822E-04 |
|---|
| alcohol catabolic process | GO:0046164 |  | 3.49E-06 | 1.421E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 3.914E-06 | 1.366E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 3.914E-06 | 1.195E-03 |
|---|
| sperm motility | GO:0030317 |  | 4.923E-06 | 1.336E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 7.429E-06 | 1.815E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.066E-05 | 2.367E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.166E-08 | 5.212E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 6.15E-07 | 7.399E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.001E-06 | 8.03E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.201E-06 | 7.223E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 2.436E-06 | 1.172E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 4.308E-06 | 1.728E-03 |
|---|
| sperm motility | GO:0030317 |  | 4.891E-06 | 1.681E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 4.891E-06 | 1.471E-03 |
|---|
| alcohol catabolic process | GO:0046164 |  | 5.235E-06 | 1.4E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.048E-05 | 2.522E-03 |
|---|
| negative regulation of adenylate cyclase activity | GO:0007194 |  | 1.503E-05 | 3.288E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.531E-08 | 3.741E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.351E-07 | 5.315E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.355E-07 | 5.175E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 8.499E-07 | 5.191E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 2.01E-06 | 9.822E-04 |
|---|
| alcohol catabolic process | GO:0046164 |  | 3.49E-06 | 1.421E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 3.914E-06 | 1.366E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 3.914E-06 | 1.195E-03 |
|---|
| sperm motility | GO:0030317 |  | 4.923E-06 | 1.336E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 7.429E-06 | 1.815E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 1.066E-05 | 2.367E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.166E-08 | 5.212E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 6.15E-07 | 7.399E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.001E-06 | 8.03E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.201E-06 | 7.223E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 2.436E-06 | 1.172E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 4.308E-06 | 1.728E-03 |
|---|
| sperm motility | GO:0030317 |  | 4.891E-06 | 1.681E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 4.891E-06 | 1.471E-03 |
|---|
| alcohol catabolic process | GO:0046164 |  | 5.235E-06 | 1.4E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.048E-05 | 2.522E-03 |
|---|
| negative regulation of adenylate cyclase activity | GO:0007194 |  | 1.503E-05 | 3.288E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 7.119E-08 | 2.542E-04 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 2.093E-07 | 3.737E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.703E-07 | 3.218E-04 |
|---|
| site of polarized growth | GO:0030427 |  | 1.938E-06 | 1.73E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.874E-05 | 0.01338717 |
|---|
| mitochondrial respiratory chain complex assembly | GO:0033108 |  | 1.874E-05 | 0.01115597 |
|---|
| hemopoietic progenitor cell differentiation | GO:0002244 |  | 1.874E-05 | 9.562E-03 |
|---|
| establishment of apical/basal cell polarity | GO:0035089 |  | 1.874E-05 | 8.367E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.878E-05 | 7.452E-03 |
|---|
| response to UV | GO:0009411 |  | 2.275E-05 | 8.123E-03 |
|---|
| eye photoreceptor cell differentiation | GO:0001754 |  | 2.622E-05 | 8.513E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.862E-07 | 6.793E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.103E-06 | 2.013E-03 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.203E-06 | 1.463E-03 |
|---|
| site of polarized growth | GO:0030427 |  | 7.514E-06 | 6.852E-03 |
|---|
| hemopoietic progenitor cell differentiation | GO:0002244 |  | 2.311E-05 | 0.01685772 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 2.311E-05 | 0.0140481 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.462E-05 | 0.01804448 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.957E-05 | 0.01804217 |
|---|
| response to UV | GO:0009411 |  | 4.54E-05 | 0.01840195 |
|---|
| mitochondrial respiratory chain complex assembly | GO:0033108 |  | 4.843E-05 | 0.01766659 |
|---|
| establishment or maintenance of epithelial cell apical/basal polarity | GO:0045197 |  | 4.843E-05 | 0.01606054 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 2.166E-08 | 5.212E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 6.15E-07 | 7.399E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.001E-06 | 8.03E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 1.201E-06 | 7.223E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 2.436E-06 | 1.172E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 4.308E-06 | 1.728E-03 |
|---|
| sperm motility | GO:0030317 |  | 4.891E-06 | 1.681E-03 |
|---|
| osteoblast differentiation | GO:0001649 |  | 4.891E-06 | 1.471E-03 |
|---|
| alcohol catabolic process | GO:0046164 |  | 5.235E-06 | 1.4E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.048E-05 | 2.522E-03 |
|---|
| negative regulation of adenylate cyclase activity | GO:0007194 |  | 1.503E-05 | 3.288E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 6.703E-08 | 1.542E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.097E-06 | 1.262E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.793E-06 | 2.141E-03 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.699E-06 | 2.127E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 7.491E-06 | 3.446E-03 |
|---|
| sperm motility | GO:0030317 |  | 1.323E-05 | 5.071E-03 |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.698E-05 | 5.581E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 2.372E-05 | 6.821E-03 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 3.08E-05 | 7.871E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 3.208E-05 | 7.377E-03 |
|---|
| hemopoietic progenitor cell differentiation | GO:0002244 |  | 3.658E-05 | 7.649E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 6.703E-08 | 1.542E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.097E-06 | 1.262E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.793E-06 | 2.141E-03 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.699E-06 | 2.127E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 7.491E-06 | 3.446E-03 |
|---|
| sperm motility | GO:0030317 |  | 1.323E-05 | 5.071E-03 |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.698E-05 | 5.581E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 2.372E-05 | 6.821E-03 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 3.08E-05 | 7.871E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 3.208E-05 | 7.377E-03 |
|---|
| hemopoietic progenitor cell differentiation | GO:0002244 |  | 3.658E-05 | 7.649E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 6.703E-08 | 1.542E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.097E-06 | 1.262E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.793E-06 | 2.141E-03 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.699E-06 | 2.127E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 7.491E-06 | 3.446E-03 |
|---|
| sperm motility | GO:0030317 |  | 1.323E-05 | 5.071E-03 |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.698E-05 | 5.581E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 2.372E-05 | 6.821E-03 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 3.08E-05 | 7.871E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 3.208E-05 | 7.377E-03 |
|---|
| hemopoietic progenitor cell differentiation | GO:0002244 |  | 3.658E-05 | 7.649E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 6.703E-08 | 1.542E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.097E-06 | 1.262E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.793E-06 | 2.141E-03 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.699E-06 | 2.127E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 7.491E-06 | 3.446E-03 |
|---|
| sperm motility | GO:0030317 |  | 1.323E-05 | 5.071E-03 |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.698E-05 | 5.581E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 2.372E-05 | 6.821E-03 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 3.08E-05 | 7.871E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 3.208E-05 | 7.377E-03 |
|---|
| hemopoietic progenitor cell differentiation | GO:0002244 |  | 3.658E-05 | 7.649E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 4.981E-08 | 1.182E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 1.087E-06 | 1.289E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.213E-06 | 1.751E-03 |
|---|
| fructose metabolic process | GO:0006000 |  | 3.338E-06 | 1.98E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 7.515E-06 | 3.566E-03 |
|---|
| sperm motility | GO:0030317 |  | 1.263E-05 | 4.994E-03 |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.54E-05 | 5.222E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 2.17E-05 | 6.436E-03 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 2.795E-05 | 7.37E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 2.878E-05 | 6.83E-03 |
|---|
| establishment of apical/basal cell polarity | GO:0035089 |  | 2.999E-05 | 6.469E-03 |
|---|



