Study VanDeVijver-ER-pos
Study informations
14 subnetworks in total page | file
272 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 88
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.5297 | 0.000e+00 | 0.000e+00 | 2.260e-04 | 0.000e+00 |
|---|
| IPC | 0.4541 | 1.130e-01 | 9.378e-02 | 9.580e-01 | 1.015e-02 |
|---|
| Loi | 0.5112 | 0.000e+00 | 0.000e+00 | 2.443e-03 | 0.000e+00 |
|---|
| Schmidt | 0.2888 | 1.618e-02 | 4.710e-03 | 3.177e-01 | 2.421e-05 |
|---|
| Wang | 0.3373 | 1.390e-04 | 4.000e-06 | 5.294e-02 | 2.943e-11 |
|---|
Expression data for subnetwork 88 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
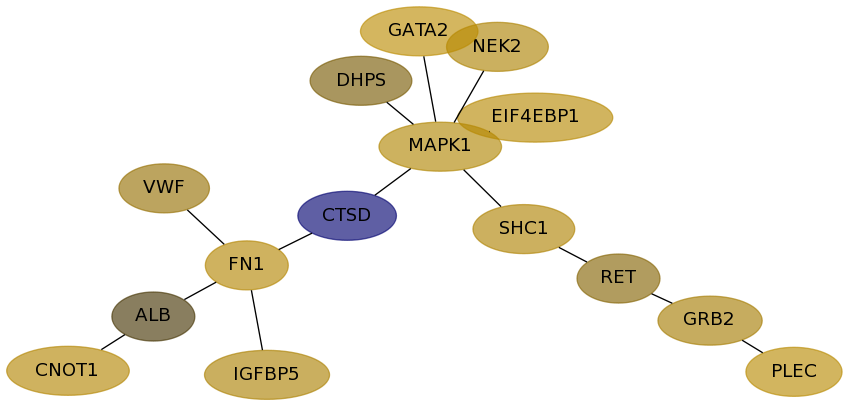
Score for each gene in subnetwork 88 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| GRB2 |   | 7 | 1 | 80 | 80 | 0.121 | 0.037 | 0.087 | 0.182 | -0.037 |
|---|
| SHC1 |   | 4 | 6 | 167 | 167 | 0.149 | -0.137 | 0.092 | -0.200 | 0.113 |
|---|
| FN1 |   | 3 | 13 | 80 | 84 | 0.167 | 0.154 | 0.006 | -0.056 | 0.137 |
|---|
| CNOT1 |   | 1 | 59 | 190 | 190 | 0.165 | 0.064 | 0.095 | 0.027 | 0.087 |
|---|
| ALB |   | 5 | 4 | 80 | 81 | 0.012 | -0.085 | 0.115 | -0.022 | -0.027 |
|---|
| GATA2 |   | 4 | 6 | 80 | 82 | 0.201 | -0.014 | 0.121 | 0.080 | -0.026 |
|---|
| DHPS |   | 1 | 59 | 190 | 190 | 0.040 | -0.067 | 0.217 | -0.110 | -0.043 |
|---|
| EIF4EBP1 |   | 2 | 27 | 80 | 87 | 0.179 | -0.044 | 0.171 | 0.066 | 0.132 |
|---|
| IGFBP5 |   | 4 | 6 | 1 | 3 | 0.142 | 0.011 | 0.158 | 0.087 | -0.000 |
|---|
| RET |   | 4 | 6 | 167 | 167 | 0.055 | -0.063 | 0.119 | 0.041 | 0.006 |
|---|
| NEK2 |   | 3 | 13 | 110 | 111 | 0.147 | 0.147 | 0.232 | 0.245 | 0.336 |
|---|
| VWF |   | 3 | 13 | 36 | 37 | 0.081 | -0.060 | -0.038 | -0.112 | -0.069 |
|---|
| PLEC |   | 1 | 59 | 190 | 190 | 0.188 | -0.132 | 0.079 | 0.006 | 0.038 |
|---|
| CTSD |   | 1 | 59 | 190 | 190 | -0.034 | 0.007 | 0.131 | 0.121 | -0.051 |
|---|
| MAPK1 |   | 5 | 4 | 110 | 110 | 0.157 | -0.023 | -0.125 | -0.115 | 0.092 |
|---|
GO Enrichment output for subnetwork 88 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of translation | GO:0017148 |  | 2.515E-06 | 5.786E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.154E-05 | 0.01327623 |
|---|
| placenta development | GO:0001890 |  | 1.451E-05 | 0.01112623 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 2.464E-05 | 0.01416691 |
|---|
| peptidyl-lysine modification to hypusine | GO:0008612 |  | 2.838E-05 | 0.01305352 |
|---|
| enteric nervous system development | GO:0048484 |  | 2.838E-05 | 0.01087794 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 4.252E-05 | 0.01397099 |
|---|
| interaction with symbiont | GO:0051702 |  | 4.252E-05 | 0.01222461 |
|---|
| response to exogenous dsRNA | GO:0043330 |  | 4.252E-05 | 0.01086632 |
|---|
| regulation of smooth muscle cell migration | GO:0014910 |  | 5.947E-05 | 0.01367695 |
|---|
| lipopolysaccharide-mediated signaling pathway | GO:0031663 |  | 5.947E-05 | 0.01243359 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of translation | GO:0017148 |  | 2.431E-06 | 5.94E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 6.935E-06 | 8.472E-03 |
|---|
| placenta development | GO:0001890 |  | 1.377E-05 | 0.01121027 |
|---|
| spermidine metabolic process | GO:0008216 |  | 1.707E-05 | 0.01042554 |
|---|
| peptidyl-lysine modification to hypusine | GO:0008612 |  | 1.707E-05 | 8.34E-03 |
|---|
| enteric nervous system development | GO:0048484 |  | 1.707E-05 | 6.95E-03 |
|---|
| interaction with symbiont | GO:0051702 |  | 2.558E-05 | 8.929E-03 |
|---|
| response to exogenous dsRNA | GO:0043330 |  | 2.558E-05 | 7.813E-03 |
|---|
| lipopolysaccharide-mediated signaling pathway | GO:0031663 |  | 3.579E-05 | 9.714E-03 |
|---|
| negative regulation of smooth muscle cell proliferation | GO:0048662 |  | 4.768E-05 | 0.01164708 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 4.768E-05 | 0.01058825 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of translation | GO:0017148 |  | 2.081E-06 | 5.008E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 5.308E-06 | 6.386E-03 |
|---|
| placenta development | GO:0001890 |  | 1.079E-05 | 8.656E-03 |
|---|
| spermidine metabolic process | GO:0008216 |  | 1.546E-05 | 9.302E-03 |
|---|
| peptidyl-lysine modification to hypusine | GO:0008612 |  | 1.546E-05 | 7.442E-03 |
|---|
| enteric nervous system development | GO:0048484 |  | 1.546E-05 | 6.201E-03 |
|---|
| interaction with symbiont | GO:0051702 |  | 2.318E-05 | 7.967E-03 |
|---|
| response to exogenous dsRNA | GO:0043330 |  | 2.318E-05 | 6.971E-03 |
|---|
| lipopolysaccharide-mediated signaling pathway | GO:0031663 |  | 3.242E-05 | 8.668E-03 |
|---|
| cell maturation | GO:0048469 |  | 4.156E-05 | 9.999E-03 |
|---|
| negative regulation of smooth muscle cell proliferation | GO:0048662 |  | 4.32E-05 | 9.448E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of translation | GO:0017148 |  | 2.515E-06 | 5.786E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.154E-05 | 0.01327623 |
|---|
| placenta development | GO:0001890 |  | 1.451E-05 | 0.01112623 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 2.464E-05 | 0.01416691 |
|---|
| peptidyl-lysine modification to hypusine | GO:0008612 |  | 2.838E-05 | 0.01305352 |
|---|
| enteric nervous system development | GO:0048484 |  | 2.838E-05 | 0.01087794 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 4.252E-05 | 0.01397099 |
|---|
| interaction with symbiont | GO:0051702 |  | 4.252E-05 | 0.01222461 |
|---|
| response to exogenous dsRNA | GO:0043330 |  | 4.252E-05 | 0.01086632 |
|---|
| regulation of smooth muscle cell migration | GO:0014910 |  | 5.947E-05 | 0.01367695 |
|---|
| lipopolysaccharide-mediated signaling pathway | GO:0031663 |  | 5.947E-05 | 0.01243359 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of translation | GO:0017148 |  | 2.515E-06 | 5.786E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.154E-05 | 0.01327623 |
|---|
| placenta development | GO:0001890 |  | 1.451E-05 | 0.01112623 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 2.464E-05 | 0.01416691 |
|---|
| peptidyl-lysine modification to hypusine | GO:0008612 |  | 2.838E-05 | 0.01305352 |
|---|
| enteric nervous system development | GO:0048484 |  | 2.838E-05 | 0.01087794 |
|---|
| negative regulation of translational initiation | GO:0045947 |  | 4.252E-05 | 0.01397099 |
|---|
| interaction with symbiont | GO:0051702 |  | 4.252E-05 | 0.01222461 |
|---|
| response to exogenous dsRNA | GO:0043330 |  | 4.252E-05 | 0.01086632 |
|---|
| regulation of smooth muscle cell migration | GO:0014910 |  | 5.947E-05 | 0.01367695 |
|---|
| lipopolysaccharide-mediated signaling pathway | GO:0031663 |  | 5.947E-05 | 0.01243359 |
|---|


