Study VanDeVijver-ER-pos
Study informations
14 subnetworks in total page | file
272 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 55
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.5171 | 0.000e+00 | 0.000e+00 | 5.210e-04 | 0.000e+00 |
|---|
| IPC | 0.3586 | 2.593e-01 | 2.606e-01 | 9.688e-01 | 6.547e-02 |
|---|
| Loi | 0.5320 | 0.000e+00 | 0.000e+00 | 1.490e-03 | 0.000e+00 |
|---|
| Schmidt | 0.3219 | 4.990e-03 | 9.530e-04 | 1.521e-01 | 7.234e-07 |
|---|
| Wang | 0.2882 | 2.177e-03 | 1.530e-04 | 2.447e-01 | 8.150e-08 |
|---|
Expression data for subnetwork 55 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
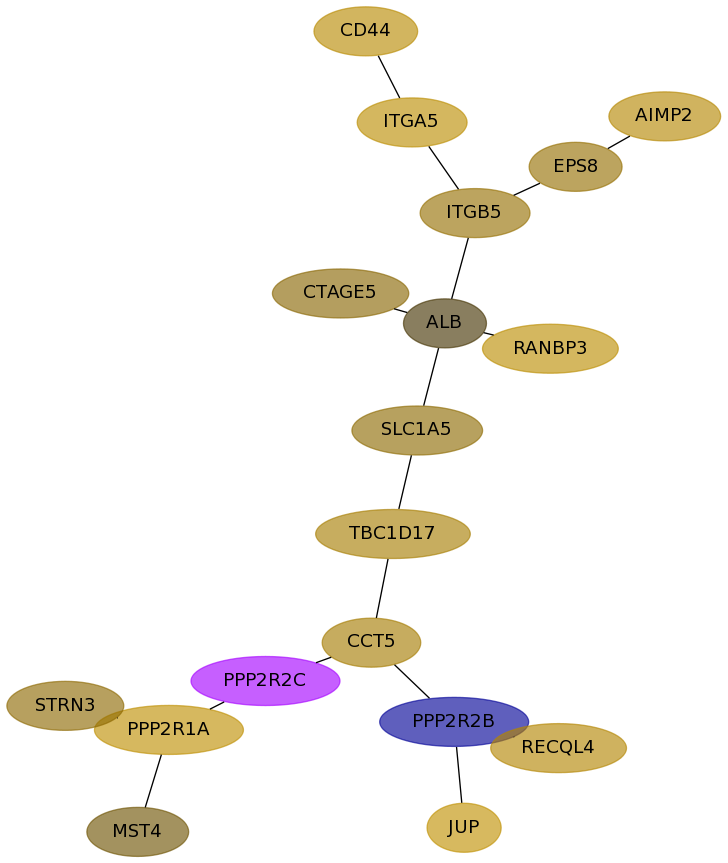
Score for each gene in subnetwork 55 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| JUP |   | 1 | 59 | 264 | 264 | 0.226 | 0.058 | 0.056 | 0.215 | -0.027 |
|---|
| EPS8 |   | 1 | 59 | 264 | 264 | 0.083 | 0.034 | 0.125 | 0.002 | 0.051 |
|---|
| ITGA5 |   | 3 | 13 | 36 | 37 | 0.204 | -0.033 | -0.023 | -0.155 | 0.135 |
|---|
| RANBP3 |   | 2 | 27 | 194 | 194 | 0.206 | 0.033 | -0.052 | -0.043 | -0.041 |
|---|
| ITGB5 |   | 1 | 59 | 264 | 264 | 0.081 | 0.045 | -0.012 | -0.093 | -0.026 |
|---|
| CCT5 |   | 2 | 27 | 65 | 65 | 0.122 | 0.132 | 0.196 | 0.122 | 0.139 |
|---|
| RECQL4 |   | 1 | 59 | 264 | 264 | 0.166 | 0.046 | 0.140 | 0.282 | 0.215 |
|---|
| TBC1D17 |   | 2 | 27 | 65 | 65 | 0.126 | -0.165 | 0.100 | -0.037 | -0.031 |
|---|
| PPP2R1A |   | 2 | 27 | 65 | 65 | 0.215 | -0.149 | 0.172 | -0.047 | -0.085 |
|---|
| ALB |   | 5 | 4 | 80 | 81 | 0.012 | -0.085 | 0.115 | -0.022 | -0.027 |
|---|
| MST4 |   | 2 | 27 | 65 | 65 | 0.033 | 0.069 | 0.222 | 0.226 | 0.070 |
|---|
| CTAGE5 |   | 1 | 59 | 264 | 264 | 0.060 | 0.076 | -0.066 | 0.037 | 0.120 |
|---|
| PPP2R2B |   | 1 | 59 | 264 | 264 | -0.086 | -0.027 | -0.083 | -0.077 | -0.091 |
|---|
| PPP2R2C |   | 2 | 27 | 65 | 65 | undef | -0.085 | 0.177 | undef | undef |
|---|
| CD44 |   | 3 | 13 | 36 | 37 | 0.186 | 0.054 | -0.053 | -0.293 | -0.279 |
|---|
| SLC1A5 |   | 1 | 59 | 264 | 264 | 0.068 | -0.093 | 0.179 | 0.028 | -0.141 |
|---|
| AIMP2 |   | 1 | 59 | 264 | 264 | 0.172 | 0.017 | 0.160 | 0.157 | 0.070 |
|---|
| STRN3 |   | 1 | 59 | 264 | 264 | 0.067 | 0.093 | 0.079 | -0.025 | 0.220 |
|---|
GO Enrichment output for subnetwork 55 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 2.649E-05 | 0.06092342 |
|---|
| negative regulation of peptidyl-tyrosine phosphorylation | GO:0050732 |  | 2.649E-05 | 0.03046171 |
|---|
| wound healing. spreading of epidermal cells | GO:0035313 |  | 3.969E-05 | 0.03043035 |
|---|
| interaction with symbiont | GO:0051702 |  | 3.969E-05 | 0.02282276 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 5.551E-05 | 0.02553518 |
|---|
| integrin-mediated signaling pathway | GO:0007229 |  | 7.183E-05 | 0.02753571 |
|---|
| mitochondrion localization | GO:0051646 |  | 7.394E-05 | 0.02429419 |
|---|
| cell-matrix adhesion | GO:0007160 |  | 1.385E-04 | 0.03980585 |
|---|
| dicarboxylic acid transport | GO:0006835 |  | 1.736E-04 | 0.04435629 |
|---|
| neutral amino acid transport | GO:0015804 |  | 1.736E-04 | 0.03992066 |
|---|
| cell-substrate adhesion | GO:0031589 |  | 1.751E-04 | 0.03660402 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 1.563E-05 | 0.03819593 |
|---|
| negative regulation of peptidyl-tyrosine phosphorylation | GO:0050732 |  | 1.563E-05 | 0.01909796 |
|---|
| interaction with symbiont | GO:0051702 |  | 2.343E-05 | 0.01908256 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 3.278E-05 | 0.02002054 |
|---|
| wound healing. spreading of epidermal cells | GO:0035313 |  | 3.278E-05 | 0.01601643 |
|---|
| mitochondrion localization | GO:0051646 |  | 5.61E-05 | 0.02284374 |
|---|
| integrin-mediated signaling pathway | GO:0007229 |  | 7.485E-05 | 0.02612123 |
|---|
| cell-matrix adhesion | GO:0007160 |  | 1.25E-04 | 0.03815807 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 1.412E-04 | 0.03834104 |
|---|
| neutral amino acid transport | GO:0015804 |  | 1.412E-04 | 0.03450694 |
|---|
| cell-substrate adhesion | GO:0031589 |  | 1.566E-04 | 0.03477459 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 1.63E-05 | 0.03921636 |
|---|
| negative regulation of peptidyl-tyrosine phosphorylation | GO:0050732 |  | 1.63E-05 | 0.01960818 |
|---|
| interaction with symbiont | GO:0051702 |  | 2.443E-05 | 0.01959212 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 3.417E-05 | 0.02055489 |
|---|
| wound healing. spreading of epidermal cells | GO:0035313 |  | 3.417E-05 | 0.01644391 |
|---|
| mitochondrion localization | GO:0051646 |  | 5.849E-05 | 0.02345285 |
|---|
| integrin-mediated signaling pathway | GO:0007229 |  | 6.207E-05 | 0.0213336 |
|---|
| cell-matrix adhesion | GO:0007160 |  | 1.271E-04 | 0.03822201 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 1.472E-04 | 0.03936091 |
|---|
| neutral amino acid transport | GO:0015804 |  | 1.472E-04 | 0.03542482 |
|---|
| cell-substrate adhesion | GO:0031589 |  | 1.597E-04 | 0.03493539 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 2.649E-05 | 0.06092342 |
|---|
| negative regulation of peptidyl-tyrosine phosphorylation | GO:0050732 |  | 2.649E-05 | 0.03046171 |
|---|
| wound healing. spreading of epidermal cells | GO:0035313 |  | 3.969E-05 | 0.03043035 |
|---|
| interaction with symbiont | GO:0051702 |  | 3.969E-05 | 0.02282276 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 5.551E-05 | 0.02553518 |
|---|
| integrin-mediated signaling pathway | GO:0007229 |  | 7.183E-05 | 0.02753571 |
|---|
| mitochondrion localization | GO:0051646 |  | 7.394E-05 | 0.02429419 |
|---|
| cell-matrix adhesion | GO:0007160 |  | 1.385E-04 | 0.03980585 |
|---|
| dicarboxylic acid transport | GO:0006835 |  | 1.736E-04 | 0.04435629 |
|---|
| neutral amino acid transport | GO:0015804 |  | 1.736E-04 | 0.03992066 |
|---|
| cell-substrate adhesion | GO:0031589 |  | 1.751E-04 | 0.03660402 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 2.649E-05 | 0.06092342 |
|---|
| negative regulation of peptidyl-tyrosine phosphorylation | GO:0050732 |  | 2.649E-05 | 0.03046171 |
|---|
| wound healing. spreading of epidermal cells | GO:0035313 |  | 3.969E-05 | 0.03043035 |
|---|
| interaction with symbiont | GO:0051702 |  | 3.969E-05 | 0.02282276 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 5.551E-05 | 0.02553518 |
|---|
| integrin-mediated signaling pathway | GO:0007229 |  | 7.183E-05 | 0.02753571 |
|---|
| mitochondrion localization | GO:0051646 |  | 7.394E-05 | 0.02429419 |
|---|
| cell-matrix adhesion | GO:0007160 |  | 1.385E-04 | 0.03980585 |
|---|
| dicarboxylic acid transport | GO:0006835 |  | 1.736E-04 | 0.04435629 |
|---|
| neutral amino acid transport | GO:0015804 |  | 1.736E-04 | 0.03992066 |
|---|
| cell-substrate adhesion | GO:0031589 |  | 1.751E-04 | 0.03660402 |
|---|


