Study VanDeVijver-ER-pos
Study informations
14 subnetworks in total page | file
272 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 103
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.5314 | 0.000e+00 | 0.000e+00 | 2.000e-04 | 0.000e+00 |
|---|
| IPC | 0.4926 | 7.326e-02 | 5.484e-02 | 9.541e-01 | 3.833e-03 |
|---|
| Loi | 0.5102 | 0.000e+00 | 0.000e+00 | 2.496e-03 | 0.000e+00 |
|---|
| Schmidt | 0.2747 | 2.518e-02 | 8.601e-03 | 4.043e-01 | 8.758e-05 |
|---|
| Wang | 0.3344 | 1.660e-04 | 4.000e-06 | 5.914e-02 | 3.927e-11 |
|---|
Expression data for subnetwork 103 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
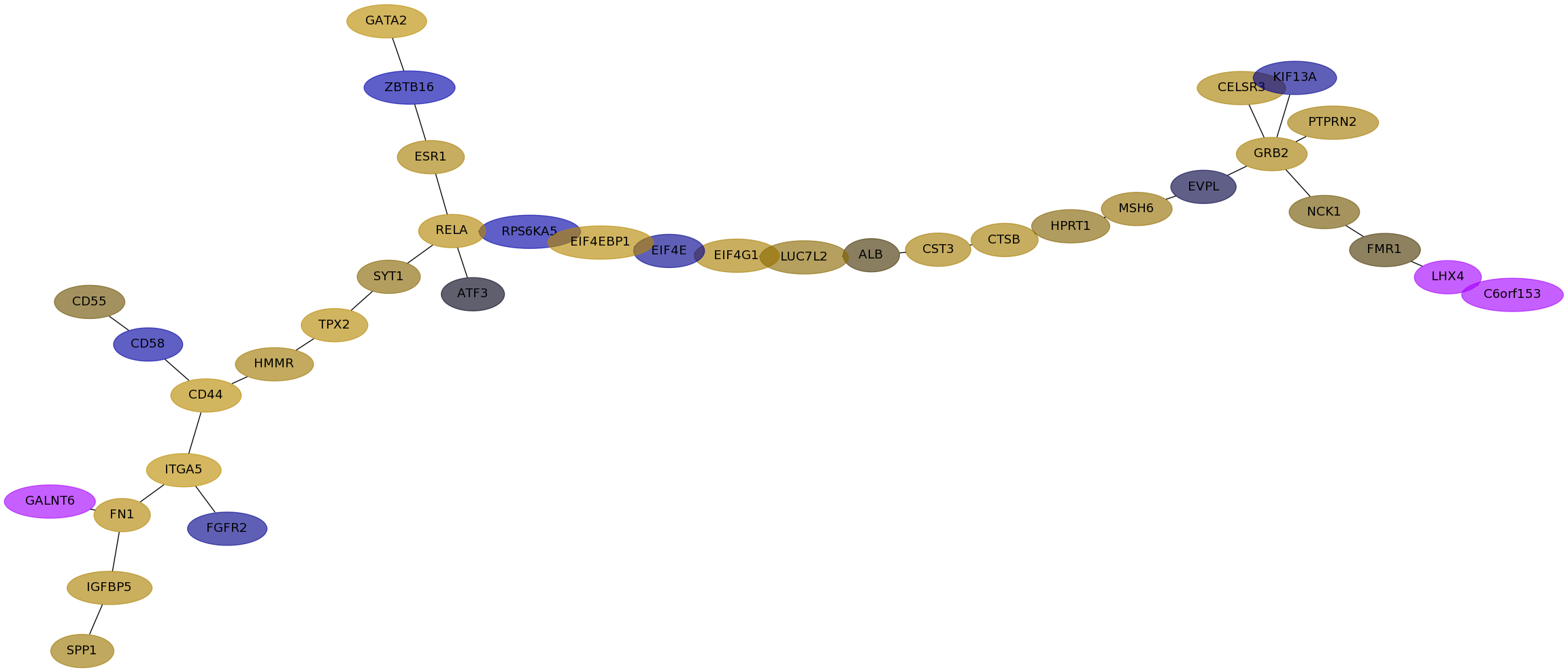
Score for each gene in subnetwork 103 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| FN1 |   | 3 | 13 | 80 | 84 | 0.167 | 0.154 | 0.006 | -0.056 | 0.137 |
|---|
| CELSR3 |   | 1 | 59 | 80 | 90 | 0.131 | 0.165 | 0.105 | 0.098 | 0.076 |
|---|
| GALNT6 |   | 1 | 59 | 80 | 90 | undef | 0.127 | 0.096 | undef | undef |
|---|
| CTSB |   | 1 | 59 | 80 | 90 | 0.122 | 0.224 | 0.079 | 0.044 | -0.020 |
|---|
| TPX2 |   | 1 | 59 | 80 | 90 | 0.182 | 0.129 | 0.156 | 0.216 | 0.145 |
|---|
| EVPL |   | 3 | 13 | 80 | 84 | -0.011 | 0.035 | 0.178 | 0.232 | -0.047 |
|---|
| RPS6KA5 |   | 1 | 59 | 80 | 90 | -0.125 | 0.092 | -0.078 | -0.112 | -0.121 |
|---|
| C6orf153 |   | 1 | 59 | 80 | 90 | undef | 0.092 | 0.035 | undef | undef |
|---|
| CD58 |   | 1 | 59 | 80 | 90 | -0.111 | -0.069 | 0.070 | -0.084 | -0.003 |
|---|
| SPP1 |   | 3 | 13 | 36 | 37 | 0.096 | 0.208 | 0.125 | -0.007 | 0.149 |
|---|
| GATA2 |   | 4 | 6 | 80 | 82 | 0.201 | -0.014 | 0.121 | 0.080 | -0.026 |
|---|
| CST3 |   | 1 | 59 | 80 | 90 | 0.122 | -0.130 | -0.026 | -0.143 | -0.133 |
|---|
| CD44 |   | 3 | 13 | 36 | 37 | 0.186 | 0.054 | -0.053 | -0.293 | -0.279 |
|---|
| EIF4E |   | 1 | 59 | 80 | 90 | -0.071 | 0.118 | 0.048 | 0.097 | 0.142 |
|---|
| FMR1 |   | 1 | 59 | 80 | 90 | 0.014 | 0.129 | 0.098 | 0.079 | 0.003 |
|---|
| RELA |   | 1 | 59 | 80 | 90 | 0.165 | -0.071 | 0.006 | -0.078 | -0.081 |
|---|
| MSH6 |   | 1 | 59 | 80 | 90 | 0.082 | 0.100 | 0.147 | 0.133 | 0.133 |
|---|
| LUC7L2 |   | 2 | 27 | 80 | 87 | 0.065 | 0.117 | -0.095 | -0.057 | 0.224 |
|---|
| HMMR |   | 2 | 27 | 36 | 41 | 0.108 | 0.202 | 0.167 | 0.194 | 0.282 |
|---|
| CD55 |   | 1 | 59 | 80 | 90 | 0.033 | 0.120 | 0.025 | -0.054 | 0.071 |
|---|
| ZBTB16 |   | 1 | 59 | 80 | 90 | -0.138 | -0.069 | -0.038 | -0.093 | -0.168 |
|---|
| NCK1 |   | 3 | 13 | 80 | 84 | 0.037 | 0.127 | 0.044 | 0.103 | 0.055 |
|---|
| ITGA5 |   | 3 | 13 | 36 | 37 | 0.204 | -0.033 | -0.023 | -0.155 | 0.135 |
|---|
| KIF13A |   | 1 | 59 | 80 | 90 | -0.070 | 0.127 | 0.057 | 0.110 | 0.043 |
|---|
| ATF3 |   | 4 | 6 | 80 | 82 | -0.003 | -0.016 | 0.092 | -0.045 | -0.079 |
|---|
| SYT1 |   | 1 | 59 | 80 | 90 | 0.060 | 0.174 | 0.145 | 0.005 | 0.006 |
|---|
| GRB2 |   | 7 | 1 | 80 | 80 | 0.121 | 0.037 | 0.087 | 0.182 | -0.037 |
|---|
| EIF4G1 |   | 1 | 59 | 80 | 90 | 0.135 | -0.047 | 0.135 | 0.044 | -0.038 |
|---|
| HPRT1 |   | 1 | 59 | 80 | 90 | 0.054 | 0.198 | 0.185 | 0.089 | 0.235 |
|---|
| LHX4 |   | 2 | 27 | 80 | 87 | undef | 0.205 | 0.125 | undef | undef |
|---|
| FGFR2 |   | 1 | 59 | 80 | 90 | -0.063 | 0.074 | -0.003 | -0.171 | 0.065 |
|---|
| ALB |   | 5 | 4 | 80 | 81 | 0.012 | -0.085 | 0.115 | -0.022 | -0.027 |
|---|
| EIF4EBP1 |   | 2 | 27 | 80 | 87 | 0.179 | -0.044 | 0.171 | 0.066 | 0.132 |
|---|
| IGFBP5 |   | 4 | 6 | 1 | 3 | 0.142 | 0.011 | 0.158 | 0.087 | -0.000 |
|---|
| PTPRN2 |   | 1 | 59 | 80 | 90 | 0.118 | 0.064 | 0.023 | -0.044 | 0.050 |
|---|
| ESR1 |   | 6 | 2 | 1 | 1 | 0.125 | 0.032 | 0.114 | -0.064 | 0.048 |
|---|
GO Enrichment output for subnetwork 103 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of translation | GO:0006417 |  | 3.354E-07 | 7.714E-04 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 8.824E-07 | 1.015E-03 |
|---|
| respiratory burst | GO:0045730 |  | 1.428E-05 | 0.01094618 |
|---|
| negative regulation of translation | GO:0017148 |  | 4.767E-05 | 0.02741241 |
|---|
| immunoglobulin mediated immune response | GO:0016064 |  | 6.462E-05 | 0.02972305 |
|---|
| peptide cross-linking | GO:0018149 |  | 6.902E-05 | 0.02645621 |
|---|
| estrogen receptor signaling pathway | GO:0030520 |  | 6.902E-05 | 0.02267675 |
|---|
| lymphocyte mediated immunity | GO:0002449 |  | 1.766E-04 | 0.05076746 |
|---|
| adaptive immune response | GO:0002250 |  | 1.998E-04 | 0.05104824 |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 1.999E-04 | 0.04596863 |
|---|
| nucleoside salvage | GO:0043174 |  | 1.999E-04 | 0.04178967 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 3.641E-07 | 8.895E-04 |
|---|
| respiratory burst | GO:0045730 |  | 5.92E-06 | 7.231E-03 |
|---|
| immunoglobulin mediated immune response | GO:0016064 |  | 2.442E-05 | 0.01988362 |
|---|
| peptide cross-linking | GO:0018149 |  | 3.409E-05 | 0.02082224 |
|---|
| estrogen receptor signaling pathway | GO:0030520 |  | 4.001E-05 | 0.01954954 |
|---|
| negative regulation of translation | GO:0017148 |  | 4.001E-05 | 0.01629128 |
|---|
| lymphocyte mediated immunity | GO:0002449 |  | 6.906E-05 | 0.0241008 |
|---|
| adaptive immune response | GO:0002250 |  | 8.243E-05 | 0.02517075 |
|---|
| positive regulation of helicase activity | GO:0051096 |  | 1.104E-04 | 0.02997284 |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 1.104E-04 | 0.02697555 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.128E-04 | 0.02504163 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 3.753E-07 | 9.03E-04 |
|---|
| respiratory burst | GO:0045730 |  | 6.102E-06 | 7.34E-03 |
|---|
| immunoglobulin mediated immune response | GO:0016064 |  | 2.337E-05 | 0.01874119 |
|---|
| peptide cross-linking | GO:0018149 |  | 3.513E-05 | 0.0211326 |
|---|
| estrogen receptor signaling pathway | GO:0030520 |  | 3.513E-05 | 0.01690608 |
|---|
| negative regulation of translation | GO:0017148 |  | 4.123E-05 | 0.01653381 |
|---|
| lymphocyte mediated immunity | GO:0002449 |  | 6.74E-05 | 0.02316649 |
|---|
| adaptive immune response | GO:0002250 |  | 8.068E-05 | 0.02426381 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.04E-04 | 0.02779833 |
|---|
| acute inflammatory response | GO:0002526 |  | 1.069E-04 | 0.02571845 |
|---|
| positive regulation of helicase activity | GO:0051096 |  | 1.128E-04 | 0.02467907 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of translation | GO:0006417 |  | 3.354E-07 | 7.714E-04 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 8.824E-07 | 1.015E-03 |
|---|
| respiratory burst | GO:0045730 |  | 1.428E-05 | 0.01094618 |
|---|
| negative regulation of translation | GO:0017148 |  | 4.767E-05 | 0.02741241 |
|---|
| immunoglobulin mediated immune response | GO:0016064 |  | 6.462E-05 | 0.02972305 |
|---|
| peptide cross-linking | GO:0018149 |  | 6.902E-05 | 0.02645621 |
|---|
| estrogen receptor signaling pathway | GO:0030520 |  | 6.902E-05 | 0.02267675 |
|---|
| lymphocyte mediated immunity | GO:0002449 |  | 1.766E-04 | 0.05076746 |
|---|
| adaptive immune response | GO:0002250 |  | 1.998E-04 | 0.05104824 |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 1.999E-04 | 0.04596863 |
|---|
| nucleoside salvage | GO:0043174 |  | 1.999E-04 | 0.04178967 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of translation | GO:0006417 |  | 3.354E-07 | 7.714E-04 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 8.824E-07 | 1.015E-03 |
|---|
| respiratory burst | GO:0045730 |  | 1.428E-05 | 0.01094618 |
|---|
| negative regulation of translation | GO:0017148 |  | 4.767E-05 | 0.02741241 |
|---|
| immunoglobulin mediated immune response | GO:0016064 |  | 6.462E-05 | 0.02972305 |
|---|
| peptide cross-linking | GO:0018149 |  | 6.902E-05 | 0.02645621 |
|---|
| estrogen receptor signaling pathway | GO:0030520 |  | 6.902E-05 | 0.02267675 |
|---|
| lymphocyte mediated immunity | GO:0002449 |  | 1.766E-04 | 0.05076746 |
|---|
| adaptive immune response | GO:0002250 |  | 1.998E-04 | 0.05104824 |
|---|
| cell adhesion mediated by integrin | GO:0033627 |  | 1.999E-04 | 0.04596863 |
|---|
| nucleoside salvage | GO:0043174 |  | 1.999E-04 | 0.04178967 |
|---|


