Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 966
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3354 | 1.375e-03 | 2.300e-05 | 1.560e-01 | 4.933e-09 |
|---|
| IPC | 0.3665 | 3.066e-01 | 1.432e-01 | 9.268e-01 | 4.069e-02 |
|---|
| Loi | 0.4536 | 1.550e-04 | 0.000e+00 | 6.588e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5871 | 2.000e-06 | 0.000e+00 | 1.413e-01 | 0.000e+00 |
|---|
| Wang | 0.2547 | 8.486e-03 | 7.079e-02 | 2.412e-01 | 1.449e-04 |
|---|
Expression data for subnetwork 966 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
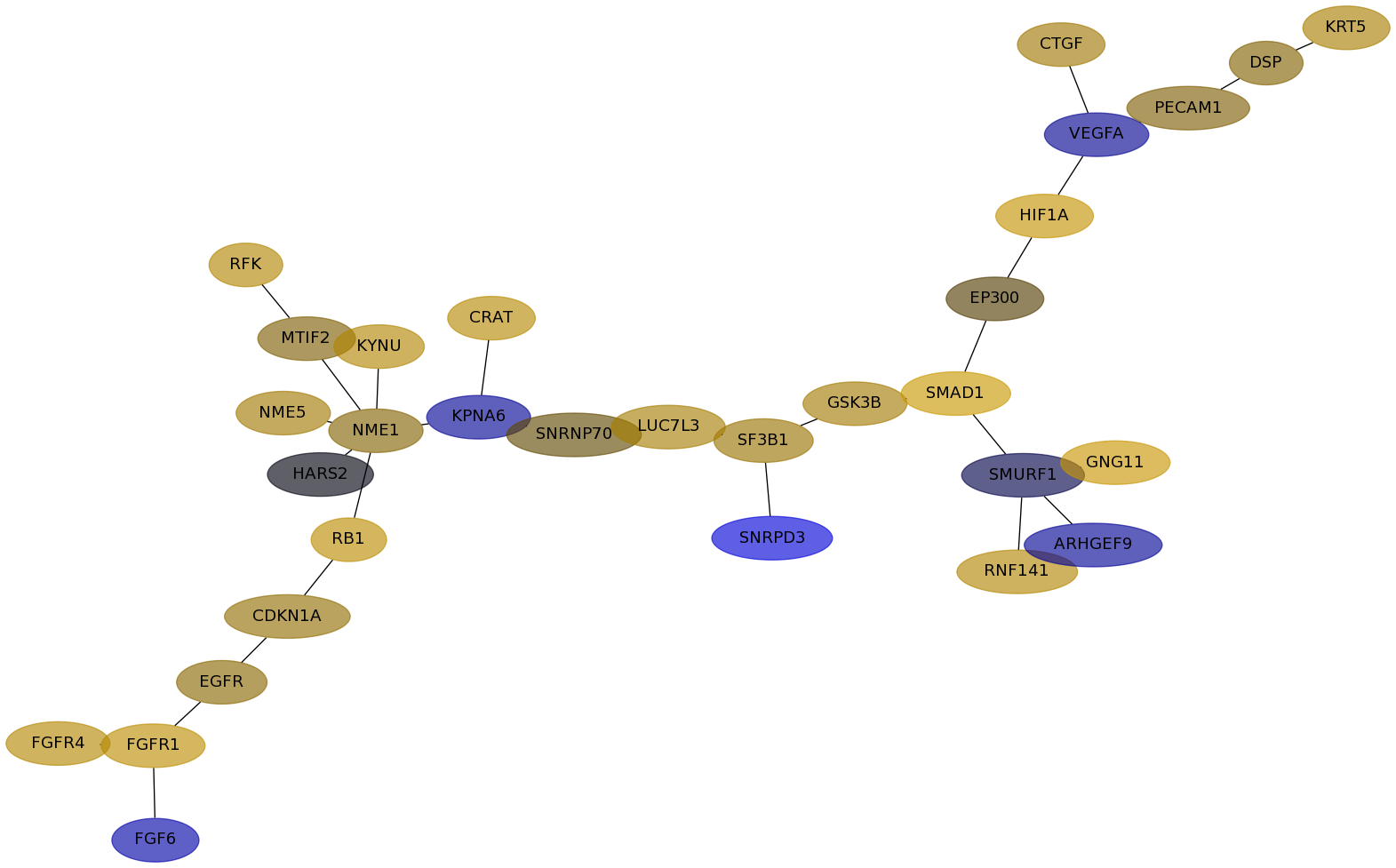
Score for each gene in subnetwork 966 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| KRT5 |   | 5 | 148 | 923 | 898 | 0.129 | -0.125 | 0.046 | 0.064 | -0.190 |
|---|
| RFK |   | 1 | 652 | 1139 | 1139 | 0.167 | 0.125 | -0.019 | 0.077 | 0.004 |
|---|
| GNG11 |   | 4 | 198 | 33 | 41 | 0.270 | 0.049 | -0.039 | 0.135 | 0.106 |
|---|
| MTIF2 |   | 4 | 198 | 492 | 499 | 0.047 | 0.013 | 0.029 | -0.088 | 0.031 |
|---|
| FGF6 |   | 2 | 391 | 963 | 966 | -0.131 | 0.125 | -0.051 | -0.159 | -0.050 |
|---|
| SF3B1 |   | 15 | 40 | 148 | 144 | 0.091 | -0.154 | 0.126 | -0.073 | -0.204 |
|---|
| NME5 |   | 1 | 652 | 1139 | 1139 | 0.111 | 0.046 | 0.054 | 0.044 | 0.038 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| PECAM1 |   | 3 | 265 | 552 | 550 | 0.046 | 0.014 | -0.161 | 0.040 | -0.106 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| SNRPD3 |   | 1 | 652 | 1139 | 1139 | -0.392 | -0.036 | -0.063 | 0.099 | -0.143 |
|---|
| DSP |   | 7 | 100 | 206 | 198 | 0.052 | 0.054 | 0.235 | -0.119 | 0.053 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| RNF141 |   | 1 | 652 | 1139 | 1139 | 0.165 | -0.127 | 0.019 | 0.079 | 0.170 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| CTGF |   | 10 | 67 | 57 | 52 | 0.106 | -0.169 | 0.181 | 0.126 | 0.209 |
|---|
| NME1 |   | 7 | 100 | 492 | 495 | 0.054 | 0.267 | 0.106 | 0.046 | -0.035 |
|---|
| HIF1A |   | 21 | 23 | 33 | 31 | 0.246 | -0.098 | 0.224 | -0.118 | 0.356 |
|---|
| CRAT |   | 1 | 652 | 1139 | 1139 | 0.183 | -0.070 | 0.139 | 0.073 | -0.012 |
|---|
| KYNU |   | 4 | 198 | 606 | 597 | 0.161 | -0.245 | 0.090 | -0.294 | 0.047 |
|---|
| LUC7L3 |   | 13 | 47 | 148 | 148 | 0.123 | 0.043 | 0.237 | 0.175 | 0.074 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| HARS2 |   | 4 | 198 | 606 | 597 | -0.002 | 0.049 | 0.360 | -0.168 | -0.199 |
|---|
| SNRNP70 |   | 3 | 265 | 994 | 971 | 0.024 | -0.129 | 0.130 | -0.059 | 0.036 |
|---|
| SMAD1 |   | 24 | 21 | 33 | 30 | 0.287 | 0.049 | 0.159 | -0.062 | 0.048 |
|---|
| SMURF1 |   | 9 | 77 | 33 | 34 | -0.013 | -0.222 | 0.236 | -0.049 | -0.006 |
|---|
| ARHGEF9 |   | 2 | 391 | 33 | 55 | -0.084 | -0.051 | 0.134 | 0.173 | 0.132 |
|---|
| KPNA6 |   | 5 | 148 | 182 | 181 | -0.082 | -0.144 | 0.128 | -0.005 | -0.108 |
|---|
GO Enrichment output for subnetwork 966 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| UTP metabolic process | GO:0046051 |  | 1.363E-08 | 3.135E-05 |
|---|
| CTP metabolic process | GO:0046036 |  | 2.447E-08 | 2.814E-05 |
|---|
| GTP metabolic process | GO:0046039 |  | 2.447E-08 | 1.876E-05 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.853E-08 | 1.641E-05 |
|---|
| pyrimidine nucleoside triphosphate biosynthetic process | GO:0009148 |  | 4.066E-08 | 1.87E-05 |
|---|
| glutathione biosynthetic process | GO:0006750 |  | 4.066E-08 | 1.559E-05 |
|---|
| pyrimidine nucleoside triphosphate metabolic process | GO:0009147 |  | 9.53E-08 | 3.131E-05 |
|---|
| pyrimidine ribonucleotide biosynthetic process | GO:0009220 |  | 1.916E-07 | 5.508E-05 |
|---|
| pyrimidine ribonucleotide metabolic process | GO:0009218 |  | 2.605E-07 | 6.658E-05 |
|---|
| peptide biosynthetic process | GO:0043043 |  | 2.605E-07 | 5.992E-05 |
|---|
| pyrimidine ribonucleoside metabolic process | GO:0046131 |  | 3.464E-07 | 7.242E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glutathione biosynthetic process | GO:0006750 |  | 7.683E-11 | 1.877E-07 |
|---|
| peptide biosynthetic process | GO:0043043 |  | 1.112E-09 | 1.359E-06 |
|---|
| UTP metabolic process | GO:0046051 |  | 5.015E-09 | 4.084E-06 |
|---|
| CTP metabolic process | GO:0046036 |  | 8.342E-09 | 5.095E-06 |
|---|
| GTP metabolic process | GO:0046039 |  | 8.342E-09 | 4.076E-06 |
|---|
| glutathione metabolic process | GO:0006749 |  | 9.232E-09 | 3.759E-06 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.113E-08 | 3.885E-06 |
|---|
| pyrimidine nucleoside triphosphate biosynthetic process | GO:0009148 |  | 1.308E-08 | 3.995E-06 |
|---|
| pyrimidine nucleoside triphosphate metabolic process | GO:0009147 |  | 2.824E-08 | 7.665E-06 |
|---|
| coenzyme biosynthetic process | GO:0009108 |  | 3.249E-08 | 7.938E-06 |
|---|
| pyrimidine ribonucleotide biosynthetic process | GO:0009220 |  | 5.37E-08 | 1.193E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glutathione biosynthetic process | GO:0006750 |  | 7.245E-11 | 1.743E-07 |
|---|
| peptide biosynthetic process | GO:0043043 |  | 1.323E-09 | 1.591E-06 |
|---|
| UTP metabolic process | GO:0046051 |  | 4.1E-09 | 3.288E-06 |
|---|
| CTP metabolic process | GO:0046036 |  | 7.364E-09 | 4.43E-06 |
|---|
| GTP metabolic process | GO:0046039 |  | 7.364E-09 | 3.544E-06 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 8.077E-09 | 3.239E-06 |
|---|
| glutathione metabolic process | GO:0006749 |  | 1.222E-08 | 4.2E-06 |
|---|
| pyrimidine nucleoside triphosphate biosynthetic process | GO:0009148 |  | 1.225E-08 | 3.683E-06 |
|---|
| pyrimidine nucleoside triphosphate metabolic process | GO:0009147 |  | 2.874E-08 | 7.684E-06 |
|---|
| coenzyme biosynthetic process | GO:0009108 |  | 4.392E-08 | 1.057E-05 |
|---|
| pyrimidine ribonucleotide biosynthetic process | GO:0009220 |  | 5.788E-08 | 1.266E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| UTP metabolic process | GO:0046051 |  | 1.363E-08 | 3.135E-05 |
|---|
| CTP metabolic process | GO:0046036 |  | 2.447E-08 | 2.814E-05 |
|---|
| GTP metabolic process | GO:0046039 |  | 2.447E-08 | 1.876E-05 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.853E-08 | 1.641E-05 |
|---|
| pyrimidine nucleoside triphosphate biosynthetic process | GO:0009148 |  | 4.066E-08 | 1.87E-05 |
|---|
| glutathione biosynthetic process | GO:0006750 |  | 4.066E-08 | 1.559E-05 |
|---|
| pyrimidine nucleoside triphosphate metabolic process | GO:0009147 |  | 9.53E-08 | 3.131E-05 |
|---|
| pyrimidine ribonucleotide biosynthetic process | GO:0009220 |  | 1.916E-07 | 5.508E-05 |
|---|
| pyrimidine ribonucleotide metabolic process | GO:0009218 |  | 2.605E-07 | 6.658E-05 |
|---|
| peptide biosynthetic process | GO:0043043 |  | 2.605E-07 | 5.992E-05 |
|---|
| pyrimidine ribonucleoside metabolic process | GO:0046131 |  | 3.464E-07 | 7.242E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| UTP metabolic process | GO:0046051 |  | 1.363E-08 | 3.135E-05 |
|---|
| CTP metabolic process | GO:0046036 |  | 2.447E-08 | 2.814E-05 |
|---|
| GTP metabolic process | GO:0046039 |  | 2.447E-08 | 1.876E-05 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.853E-08 | 1.641E-05 |
|---|
| pyrimidine nucleoside triphosphate biosynthetic process | GO:0009148 |  | 4.066E-08 | 1.87E-05 |
|---|
| glutathione biosynthetic process | GO:0006750 |  | 4.066E-08 | 1.559E-05 |
|---|
| pyrimidine nucleoside triphosphate metabolic process | GO:0009147 |  | 9.53E-08 | 3.131E-05 |
|---|
| pyrimidine ribonucleotide biosynthetic process | GO:0009220 |  | 1.916E-07 | 5.508E-05 |
|---|
| pyrimidine ribonucleotide metabolic process | GO:0009218 |  | 2.605E-07 | 6.658E-05 |
|---|
| peptide biosynthetic process | GO:0043043 |  | 2.605E-07 | 5.992E-05 |
|---|
| pyrimidine ribonucleoside metabolic process | GO:0046131 |  | 3.464E-07 | 7.242E-05 |
|---|


