Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 905
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3445 | 8.980e-04 | 1.100e-05 | 1.295e-01 | 1.279e-09 |
|---|
| IPC | 0.3404 | 3.494e-01 | 1.790e-01 | 9.358e-01 | 5.852e-02 |
|---|
| Loi | 0.4472 | 2.040e-04 | 0.000e+00 | 6.968e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5747 | 3.000e-06 | 0.000e+00 | 1.730e-01 | 0.000e+00 |
|---|
| Wang | 0.2487 | 1.073e-02 | 8.190e-02 | 2.683e-01 | 2.357e-04 |
|---|
Expression data for subnetwork 905 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
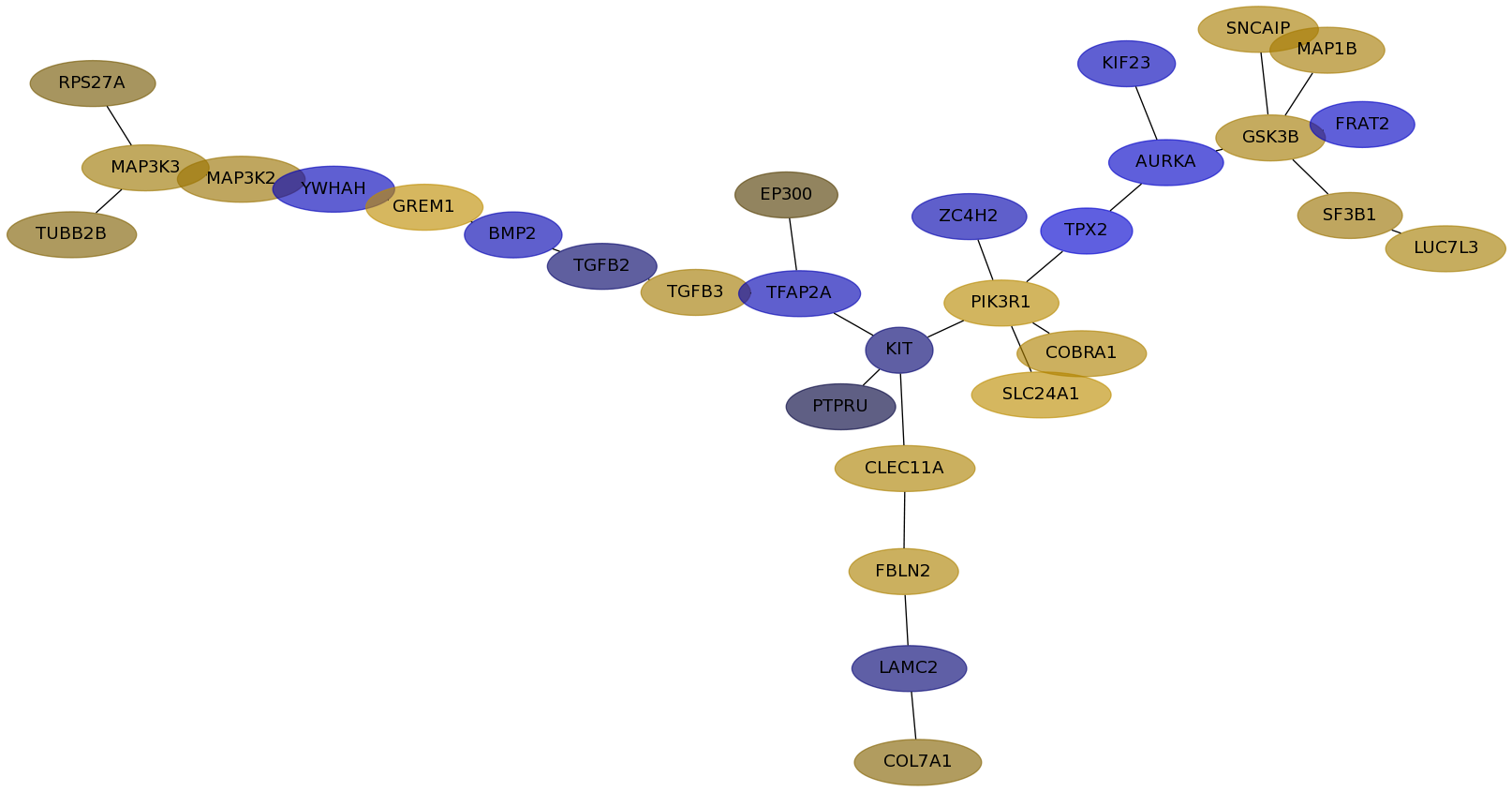
Score for each gene in subnetwork 905 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| MAP3K3 |   | 4 | 198 | 206 | 205 | 0.102 | 0.085 | 0.046 | 0.028 | 0.094 |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| GREM1 |   | 5 | 148 | 57 | 63 | 0.213 | -0.073 | 0.200 | 0.028 | 0.261 |
|---|
| CLEC11A |   | 5 | 148 | 339 | 337 | 0.146 | 0.004 | 0.353 | 0.005 | 0.272 |
|---|
| FBLN2 |   | 1 | 652 | 1436 | 1436 | 0.146 | -0.036 | 0.122 | 0.207 | -0.007 |
|---|
| PTPRU |   | 1 | 652 | 1436 | 1436 | -0.009 | 0.005 | 0.180 | 0.171 | -0.085 |
|---|
| ZC4H2 |   | 16 | 34 | 339 | 332 | -0.145 | 0.065 | 0.061 | 0.304 | 0.059 |
|---|
| SF3B1 |   | 15 | 40 | 148 | 144 | 0.091 | -0.154 | 0.126 | -0.073 | -0.204 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| TPX2 |   | 2 | 391 | 492 | 506 | -0.316 | 0.114 | -0.121 | -0.018 | -0.056 |
|---|
| KIF23 |   | 1 | 652 | 1436 | 1436 | -0.186 | 0.168 | -0.063 | 0.162 | 0.033 |
|---|
| FRAT2 |   | 2 | 391 | 1218 | 1206 | -0.244 | 0.097 | -0.042 | 0.249 | 0.048 |
|---|
| COL7A1 |   | 2 | 391 | 1135 | 1135 | 0.054 | -0.052 | 0.240 | 0.233 | 0.022 |
|---|
| TGFB2 |   | 7 | 100 | 57 | 60 | -0.028 | -0.063 | 0.244 | 0.226 | 0.015 |
|---|
| MAP1B |   | 13 | 47 | 148 | 148 | 0.118 | -0.123 | 0.263 | -0.107 | 0.100 |
|---|
| BMP2 |   | 7 | 100 | 33 | 37 | -0.155 | -0.011 | 0.097 | 0.148 | -0.069 |
|---|
| MAP3K2 |   | 2 | 391 | 552 | 554 | 0.091 | 0.115 | 0.087 | -0.066 | -0.117 |
|---|
| SLC24A1 |   | 2 | 391 | 740 | 745 | 0.209 | -0.068 | 0.109 | 0.178 | 0.056 |
|---|
| RPS27A |   | 1 | 652 | 1436 | 1436 | 0.037 | 0.050 | -0.001 | 0.202 | -0.071 |
|---|
| LUC7L3 |   | 13 | 47 | 148 | 148 | 0.123 | 0.043 | 0.237 | 0.175 | 0.074 |
|---|
| TGFB3 |   | 2 | 391 | 1003 | 998 | 0.116 | -0.190 | 0.191 | -0.088 | -0.043 |
|---|
| TFAP2A |   | 2 | 391 | 563 | 567 | -0.162 | -0.070 | 0.146 | -0.095 | -0.009 |
|---|
| AURKA |   | 1 | 652 | 1436 | 1436 | -0.261 | 0.189 | -0.176 | 0.100 | 0.050 |
|---|
| TUBB2B |   | 5 | 148 | 148 | 161 | 0.048 | -0.036 | 0.304 | 0.045 | 0.139 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| YWHAH |   | 1 | 652 | 1436 | 1436 | -0.186 | -0.082 | -0.046 | 0.001 | -0.008 |
|---|
| KIT |   | 13 | 47 | 339 | 335 | -0.034 | -0.130 | -0.091 | 0.235 | -0.059 |
|---|
| SNCAIP |   | 4 | 198 | 283 | 289 | 0.127 | -0.091 | 0.013 | 0.092 | 0.059 |
|---|
| LAMC2 |   | 2 | 391 | 1135 | 1135 | -0.037 | -0.063 | 0.307 | -0.060 | 0.055 |
|---|
| COBRA1 |   | 4 | 198 | 206 | 205 | 0.150 | 0.137 | 0.335 | 0.159 | -0.016 |
|---|
GO Enrichment output for subnetwork 905 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 5.802E-08 | 1.334E-04 |
|---|
| positive regulation of ossification | GO:0045778 |  | 2.223E-07 | 2.556E-04 |
|---|
| positive regulation of bone remodeling | GO:0046852 |  | 3.418E-07 | 2.621E-04 |
|---|
| salivary gland morphogenesis | GO:0007435 |  | 4.376E-07 | 2.516E-04 |
|---|
| SMAD protein nuclear translocation | GO:0007184 |  | 1.827E-06 | 8.404E-04 |
|---|
| salivary gland development | GO:0007431 |  | 2.605E-06 | 9.985E-04 |
|---|
| mesenchymal cell development | GO:0014031 |  | 4.068E-06 | 1.336E-03 |
|---|
| gland morphogenesis | GO:0022612 |  | 4.756E-06 | 1.367E-03 |
|---|
| exocrine system development | GO:0035272 |  | 6.17E-06 | 1.577E-03 |
|---|
| positive regulation of bone mineralization | GO:0030501 |  | 6.17E-06 | 1.419E-03 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 7.837E-06 | 1.639E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 1.962E-08 | 4.793E-05 |
|---|
| positive regulation of ossification | GO:0045778 |  | 6.911E-08 | 8.442E-05 |
|---|
| positive regulation of bone remodeling | GO:0046852 |  | 1.04E-07 | 8.471E-05 |
|---|
| salivary gland morphogenesis | GO:0007435 |  | 1.521E-07 | 9.289E-05 |
|---|
| SMAD protein nuclear translocation | GO:0007184 |  | 9.073E-07 | 4.433E-04 |
|---|
| salivary gland development | GO:0007431 |  | 9.073E-07 | 3.694E-04 |
|---|
| mesenchymal cell development | GO:0014031 |  | 1.399E-06 | 4.884E-04 |
|---|
| exocrine system development | GO:0035272 |  | 2.153E-06 | 6.575E-04 |
|---|
| gland morphogenesis | GO:0022612 |  | 2.153E-06 | 5.845E-04 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 2.736E-06 | 6.685E-04 |
|---|
| positive regulation of bone mineralization | GO:0030501 |  | 2.736E-06 | 6.077E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 3.378E-08 | 8.128E-05 |
|---|
| positive regulation of ossification | GO:0045778 |  | 1.189E-07 | 1.43E-04 |
|---|
| positive regulation of bone remodeling | GO:0046852 |  | 1.789E-07 | 1.434E-04 |
|---|
| salivary gland morphogenesis | GO:0007435 |  | 2.29E-07 | 1.378E-04 |
|---|
| SMAD protein nuclear translocation | GO:0007184 |  | 1.365E-06 | 6.57E-04 |
|---|
| salivary gland development | GO:0007431 |  | 1.365E-06 | 5.475E-04 |
|---|
| mesenchymal cell development | GO:0014031 |  | 2.166E-06 | 7.446E-04 |
|---|
| gland morphogenesis | GO:0022612 |  | 2.495E-06 | 7.503E-04 |
|---|
| exocrine system development | GO:0035272 |  | 3.238E-06 | 8.656E-04 |
|---|
| positive regulation of bone mineralization | GO:0030501 |  | 4.114E-06 | 9.899E-04 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 4.114E-06 | 8.999E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 5.802E-08 | 1.334E-04 |
|---|
| positive regulation of ossification | GO:0045778 |  | 2.223E-07 | 2.556E-04 |
|---|
| positive regulation of bone remodeling | GO:0046852 |  | 3.418E-07 | 2.621E-04 |
|---|
| salivary gland morphogenesis | GO:0007435 |  | 4.376E-07 | 2.516E-04 |
|---|
| SMAD protein nuclear translocation | GO:0007184 |  | 1.827E-06 | 8.404E-04 |
|---|
| salivary gland development | GO:0007431 |  | 2.605E-06 | 9.985E-04 |
|---|
| mesenchymal cell development | GO:0014031 |  | 4.068E-06 | 1.336E-03 |
|---|
| gland morphogenesis | GO:0022612 |  | 4.756E-06 | 1.367E-03 |
|---|
| exocrine system development | GO:0035272 |  | 6.17E-06 | 1.577E-03 |
|---|
| positive regulation of bone mineralization | GO:0030501 |  | 6.17E-06 | 1.419E-03 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 7.837E-06 | 1.639E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 5.802E-08 | 1.334E-04 |
|---|
| positive regulation of ossification | GO:0045778 |  | 2.223E-07 | 2.556E-04 |
|---|
| positive regulation of bone remodeling | GO:0046852 |  | 3.418E-07 | 2.621E-04 |
|---|
| salivary gland morphogenesis | GO:0007435 |  | 4.376E-07 | 2.516E-04 |
|---|
| SMAD protein nuclear translocation | GO:0007184 |  | 1.827E-06 | 8.404E-04 |
|---|
| salivary gland development | GO:0007431 |  | 2.605E-06 | 9.985E-04 |
|---|
| mesenchymal cell development | GO:0014031 |  | 4.068E-06 | 1.336E-03 |
|---|
| gland morphogenesis | GO:0022612 |  | 4.756E-06 | 1.367E-03 |
|---|
| exocrine system development | GO:0035272 |  | 6.17E-06 | 1.577E-03 |
|---|
| positive regulation of bone mineralization | GO:0030501 |  | 6.17E-06 | 1.419E-03 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 7.837E-06 | 1.639E-03 |
|---|


