Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 901
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3446 | 8.950e-04 | 1.100e-05 | 1.293e-01 | 1.273e-09 |
|---|
| IPC | 0.3433 | 3.447e-01 | 1.748e-01 | 9.348e-01 | 5.632e-02 |
|---|
| Loi | 0.4460 | 2.150e-04 | 0.000e+00 | 7.045e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5741 | 4.000e-06 | 0.000e+00 | 1.748e-01 | 0.000e+00 |
|---|
| Wang | 0.2491 | 1.057e-02 | 8.117e-02 | 2.666e-01 | 2.287e-04 |
|---|
Expression data for subnetwork 901 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
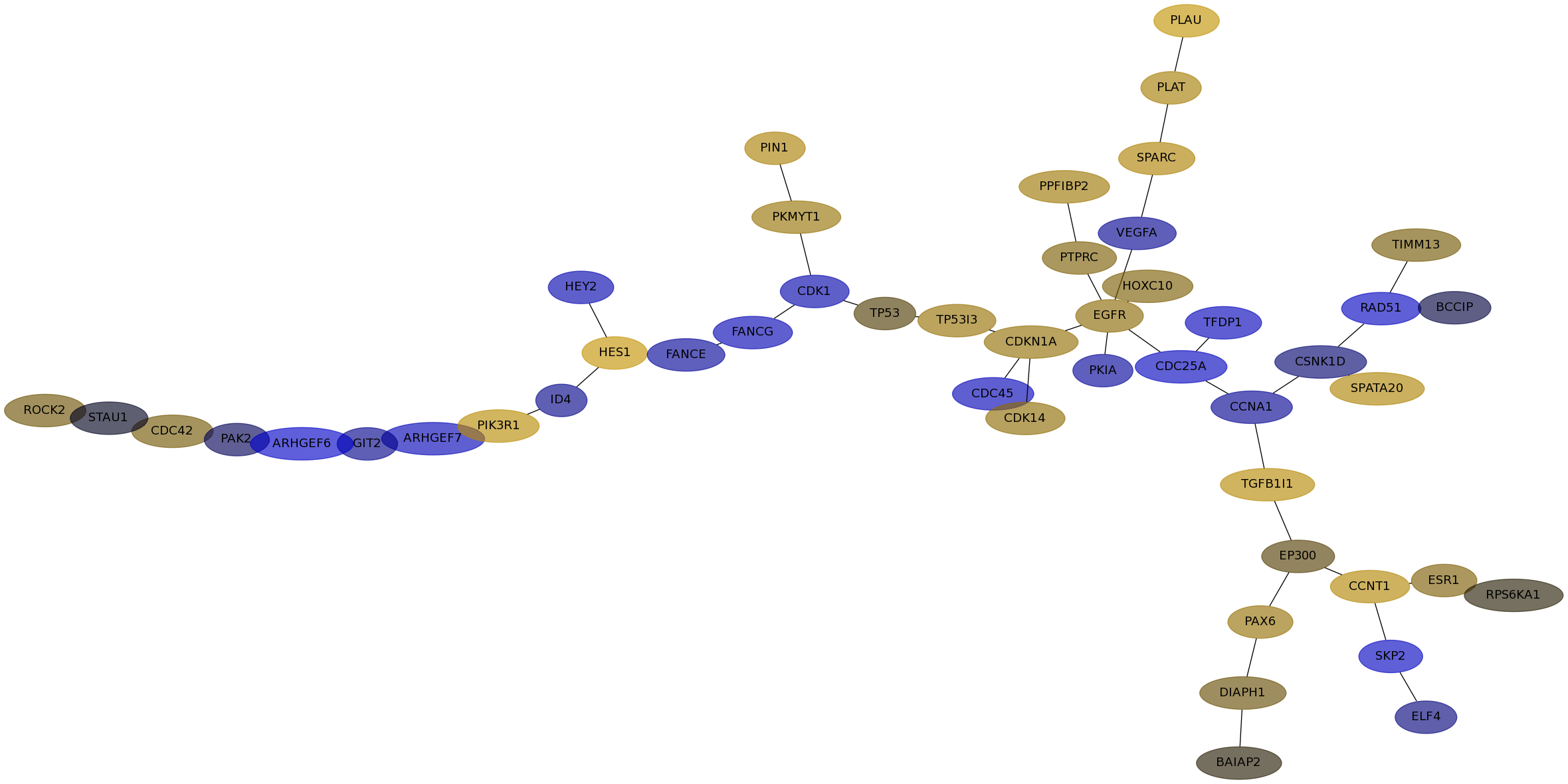
Score for each gene in subnetwork 901 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| BCCIP |   | 1 | 652 | 1422 | 1422 | -0.010 | 0.176 | -0.138 | 0.119 | -0.156 |
|---|
| PAX6 |   | 11 | 59 | 283 | 274 | 0.081 | 0.259 | -0.003 | 0.112 | 0.168 |
|---|
| ARHGEF7 |   | 12 | 55 | 492 | 475 | -0.180 | -0.074 | -0.061 | 0.208 | 0.015 |
|---|
| RPS6KA1 |   | 8 | 84 | 696 | 669 | 0.005 | -0.208 | 0.084 | -0.322 | -0.071 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| PPFIBP2 |   | 7 | 100 | 606 | 576 | 0.098 | 0.188 | 0.320 | -0.112 | 0.013 |
|---|
| ID4 |   | 13 | 47 | 444 | 430 | -0.060 | -0.179 | 0.137 | 0.241 | 0.036 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| PLAT |   | 1 | 652 | 1422 | 1422 | 0.122 | -0.096 | 0.089 | 0.090 | 0.023 |
|---|
| GIT2 |   | 3 | 265 | 584 | 573 | -0.065 | -0.191 | 0.362 | -0.164 | 0.054 |
|---|
| CDK14 |   | 2 | 391 | 1356 | 1337 | 0.067 | -0.151 | 0.011 | -0.071 | 0.107 |
|---|
| HEY2 |   | 2 | 391 | 746 | 751 | -0.147 | -0.045 | -0.021 | 0.398 | -0.058 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| PLAU |   | 6 | 120 | 444 | 438 | 0.238 | 0.187 | 0.153 | 0.141 | 0.192 |
|---|
| SPARC |   | 1 | 652 | 1422 | 1422 | 0.143 | -0.061 | 0.193 | 0.117 | 0.076 |
|---|
| PIN1 |   | 17 | 30 | 1 | 13 | 0.135 | 0.194 | 0.040 | -0.040 | -0.048 |
|---|
| PAK2 |   | 20 | 24 | 1 | 11 | -0.019 | 0.014 | 0.161 | 0.081 | 0.062 |
|---|
| ROCK2 |   | 1 | 652 | 1422 | 1422 | 0.031 | -0.178 | 0.018 | 0.278 | 0.095 |
|---|
| HOXC10 |   | 4 | 198 | 444 | 444 | 0.044 | -0.015 | 0.116 | -0.133 | -0.017 |
|---|
| SPATA20 |   | 1 | 652 | 1422 | 1422 | 0.144 | 0.016 | 0.232 | -0.103 | -0.123 |
|---|
| HES1 |   | 4 | 198 | 746 | 725 | 0.247 | 0.115 | 0.157 | 0.010 | -0.106 |
|---|
| STAU1 |   | 1 | 652 | 1422 | 1422 | -0.004 | -0.099 | 0.013 | 0.105 | 0.132 |
|---|
| CDC25A |   | 5 | 148 | 444 | 440 | -0.220 | 0.113 | -0.170 | 0.151 | -0.003 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| SKP2 |   | 14 | 42 | 148 | 146 | -0.228 | 0.092 | -0.043 | 0.237 | -0.009 |
|---|
| PKMYT1 |   | 5 | 148 | 620 | 603 | 0.085 | 0.175 | -0.143 | 0.046 | -0.070 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| FANCG |   | 1 | 652 | 1422 | 1422 | -0.170 | 0.156 | -0.281 | 0.199 | 0.147 |
|---|
| FANCE |   | 1 | 652 | 1422 | 1422 | -0.090 | 0.184 | -0.209 | 0.079 | 0.106 |
|---|
| DIAPH1 |   | 2 | 391 | 746 | 751 | 0.027 | 0.088 | 0.139 | -0.074 | 0.086 |
|---|
| ELF4 |   | 2 | 391 | 283 | 301 | -0.044 | 0.189 | 0.042 | 0.006 | 0.111 |
|---|
| CDK1 |   | 16 | 34 | 206 | 194 | -0.138 | 0.265 | -0.195 | 0.106 | 0.031 |
|---|
| TP53I3 |   | 2 | 391 | 1052 | 1044 | 0.081 | -0.065 | 0.133 | 0.123 | -0.007 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| BAIAP2 |   | 2 | 391 | 746 | 751 | 0.005 | -0.077 | -0.025 | 0.099 | -0.051 |
|---|
| PTPRC |   | 5 | 148 | 715 | 697 | 0.043 | -0.031 | -0.204 | -0.050 | -0.068 |
|---|
| CDC45 |   | 4 | 198 | 847 | 830 | -0.177 | 0.134 | -0.171 | 0.185 | -0.085 |
|---|
| TIMM13 |   | 1 | 652 | 1422 | 1422 | 0.036 | 0.091 | -0.078 | 0.164 | 0.089 |
|---|
| RAD51 |   | 4 | 198 | 696 | 684 | -0.225 | 0.146 | -0.098 | 0.135 | 0.061 |
|---|
| ARHGEF6 |   | 3 | 265 | 859 | 843 | -0.274 | -0.068 | -0.120 | 0.166 | -0.115 |
|---|
| CCNT1 |   | 18 | 26 | 33 | 32 | 0.163 | 0.008 | -0.028 | 0.162 | -0.213 |
|---|
| TFDP1 |   | 4 | 198 | 339 | 341 | -0.194 | 0.123 | -0.178 | 0.129 | -0.065 |
|---|
| CCNA1 |   | 17 | 30 | 326 | 314 | -0.078 | 0.174 | 0.092 | 0.078 | -0.098 |
|---|
| ESR1 |   | 4 | 198 | 1292 | 1277 | 0.046 | 0.026 | -0.002 | -0.053 | -0.062 |
|---|
| PKIA |   | 3 | 265 | 832 | 816 | -0.091 | -0.120 | 0.085 | 0.249 | 0.044 |
|---|
| TGFB1I1 |   | 16 | 34 | 326 | 315 | 0.176 | -0.105 | 0.229 | 0.143 | 0.298 |
|---|
| CSNK1D |   | 2 | 391 | 1198 | 1184 | -0.034 | -0.222 | -0.044 | 0.170 | 0.013 |
|---|
GO Enrichment output for subnetwork 901 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.146E-08 | 2.637E-05 |
|---|
| filopodium assembly | GO:0046847 |  | 2.736E-07 | 3.146E-04 |
|---|
| microspike assembly | GO:0030035 |  | 4.086E-07 | 3.133E-04 |
|---|
| nuclear migration | GO:0007097 |  | 1.592E-06 | 9.154E-04 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 1.592E-06 | 7.323E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.592E-06 | 6.103E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 3.171E-06 | 1.042E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.668E-06 | 1.055E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.528E-06 | 1.413E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 5.528E-06 | 1.271E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 8.809E-06 | 1.842E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.879E-09 | 4.591E-06 |
|---|
| filopodium assembly | GO:0046847 |  | 4.166E-07 | 5.088E-04 |
|---|
| muscle cell migration | GO:0014812 |  | 4.979E-07 | 4.055E-04 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 4.979E-07 | 3.041E-04 |
|---|
| microspike assembly | GO:0030035 |  | 5.34E-07 | 2.609E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.94E-07 | 3.233E-04 |
|---|
| nuclear migration | GO:0007097 |  | 9.931E-07 | 3.466E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 9.931E-07 | 3.033E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.766E-06 | 7.507E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.766E-06 | 6.756E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 4.137E-06 | 9.188E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.662E-09 | 6.404E-06 |
|---|
| filopodium assembly | GO:0046847 |  | 3.929E-07 | 4.727E-04 |
|---|
| microspike assembly | GO:0030035 |  | 5.123E-07 | 4.109E-04 |
|---|
| muscle cell migration | GO:0014812 |  | 5.83E-07 | 3.507E-04 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 5.83E-07 | 2.805E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.022E-06 | 4.098E-04 |
|---|
| nuclear migration | GO:0007097 |  | 1.163E-06 | 3.996E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.163E-06 | 3.496E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.029E-06 | 5.423E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 2.029E-06 | 4.881E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 3.237E-06 | 7.079E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.146E-08 | 2.637E-05 |
|---|
| filopodium assembly | GO:0046847 |  | 2.736E-07 | 3.146E-04 |
|---|
| microspike assembly | GO:0030035 |  | 4.086E-07 | 3.133E-04 |
|---|
| nuclear migration | GO:0007097 |  | 1.592E-06 | 9.154E-04 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 1.592E-06 | 7.323E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.592E-06 | 6.103E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 3.171E-06 | 1.042E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.668E-06 | 1.055E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.528E-06 | 1.413E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 5.528E-06 | 1.271E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 8.809E-06 | 1.842E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.146E-08 | 2.637E-05 |
|---|
| filopodium assembly | GO:0046847 |  | 2.736E-07 | 3.146E-04 |
|---|
| microspike assembly | GO:0030035 |  | 4.086E-07 | 3.133E-04 |
|---|
| nuclear migration | GO:0007097 |  | 1.592E-06 | 9.154E-04 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 1.592E-06 | 7.323E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.592E-06 | 6.103E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 3.171E-06 | 1.042E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.668E-06 | 1.055E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.528E-06 | 1.413E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 5.528E-06 | 1.271E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 8.809E-06 | 1.842E-03 |
|---|


