Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 809
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3382 | 1.211e-03 | 1.800e-05 | 1.476e-01 | 3.217e-09 |
|---|
| IPC | 0.3345 | 3.591e-01 | 1.876e-01 | 9.378e-01 | 6.316e-02 |
|---|
| Loi | 0.4502 | 1.790e-04 | 0.000e+00 | 6.784e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5665 | 5.000e-06 | 0.000e+00 | 1.962e-01 | 0.000e+00 |
|---|
| Wang | 0.2387 | 1.558e-02 | 1.032e-01 | 3.164e-01 | 5.085e-04 |
|---|
Expression data for subnetwork 809 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
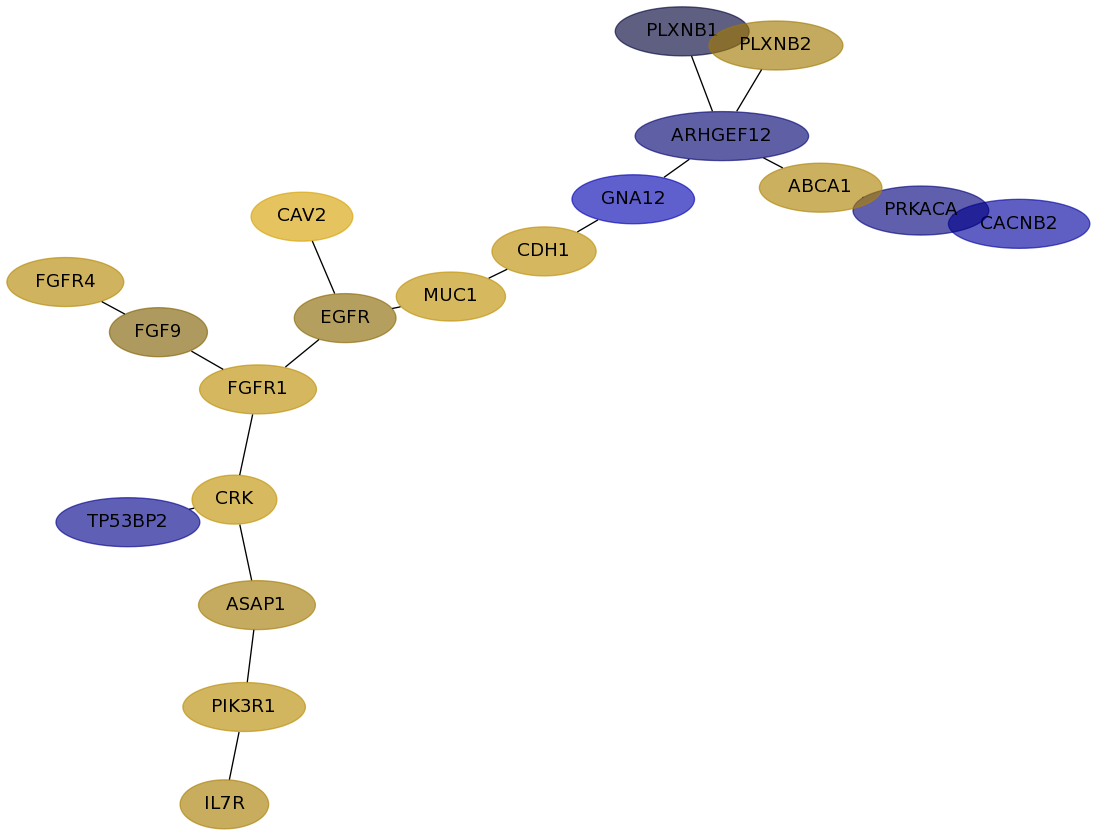
Subnetwork structure for each dataset
- Desmedt
Score for each gene in subnetwork 809 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| FGF9 |   | 5 | 148 | 262 | 265 | 0.050 | -0.096 | 0.087 | 0.014 | 0.174 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| ARHGEF12 |   | 1 | 652 | 1463 | 1463 | -0.036 | -0.190 | 0.167 | -0.018 | 0.126 |
|---|
| PRKACA |   | 1 | 652 | 1463 | 1463 | -0.047 | -0.163 | 0.181 | 0.104 | -0.144 |
|---|
| PLXNB2 |   | 1 | 652 | 1463 | 1463 | 0.109 | -0.092 | 0.308 | -0.136 | -0.062 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| PLXNB1 |   | 2 | 391 | 1232 | 1220 | -0.009 | -0.117 | 0.312 | 0.006 | -0.146 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| GNA12 |   | 1 | 652 | 1463 | 1463 | -0.160 | 0.078 | -0.009 | -0.083 | -0.027 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CRK |   | 27 | 17 | 206 | 193 | 0.232 | -0.116 | 0.106 | 0.090 | 0.017 |
|---|
| IL7R |   | 14 | 42 | 339 | 334 | 0.130 | -0.151 | -0.112 | -0.130 | -0.160 |
|---|
| ASAP1 |   | 6 | 120 | 129 | 128 | 0.114 | 0.036 | 0.070 | 0.035 | -0.056 |
|---|
| CACNB2 |   | 1 | 652 | 1463 | 1463 | -0.107 | -0.176 | 0.085 | -0.059 | -0.085 |
|---|
| TP53BP2 |   | 5 | 148 | 1 | 22 | -0.067 | 0.245 | -0.012 | 0.129 | 0.017 |
|---|
| ABCA1 |   | 1 | 652 | 1463 | 1463 | 0.144 | -0.093 | 0.037 | 0.070 | 0.043 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
GO Enrichment output for subnetwork 809 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of axonogenesis | GO:0050772 |  | 6.081E-06 | 0.01398732 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.604E-05 | 0.01844678 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.884E-05 | 0.0144414 |
|---|
| positive regulation of cell projection organization | GO:0031346 |  | 3.32E-05 | 0.01908854 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 3.32E-05 | 0.01527083 |
|---|
| regulation of cell morphogenesis | GO:0022604 |  | 4.686E-05 | 0.01796274 |
|---|
| cholesterol homeostasis | GO:0042632 |  | 5.334E-05 | 0.0175253 |
|---|
| regulation of axonogenesis | GO:0050770 |  | 5.938E-05 | 0.01707253 |
|---|
| cholesterol transport | GO:0030301 |  | 6.586E-05 | 0.01683071 |
|---|
| positive regulation of neurogenesis | GO:0050769 |  | 6.586E-05 | 0.01514764 |
|---|
| regulation of cardiac muscle cell proliferation | GO:0060043 |  | 6.739E-05 | 0.0140916 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of axonogenesis | GO:0050772 |  | 4.196E-06 | 0.01025058 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.215E-05 | 0.01484383 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.841E-05 | 0.01499023 |
|---|
| positive regulation of cell projection organization | GO:0031346 |  | 2.648E-05 | 0.0161726 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 3.298E-05 | 0.01611298 |
|---|
| regulation of cardiac muscle cell proliferation | GO:0060043 |  | 4.493E-05 | 0.01829436 |
|---|
| cytokine secretion | GO:0050663 |  | 4.493E-05 | 0.01568088 |
|---|
| regulation of phospholipase A2 activity | GO:0032429 |  | 4.493E-05 | 0.01372077 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 4.493E-05 | 0.01219624 |
|---|
| negative regulation of T cell mediated immunity | GO:0002710 |  | 4.493E-05 | 0.01097662 |
|---|
| secretory granule organization | GO:0033363 |  | 4.493E-05 | 9.979E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of axonogenesis | GO:0050772 |  | 4.488E-06 | 0.01079872 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.116E-05 | 0.01342239 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.503E-05 | 0.01205015 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.233E-05 | 0.0134344 |
|---|
| positive regulation of cell projection organization | GO:0031346 |  | 2.832E-05 | 0.01362526 |
|---|
| cholesterol homeostasis | GO:0042632 |  | 4.324E-05 | 0.01733983 |
|---|
| regulation of cell morphogenesis | GO:0022604 |  | 4.464E-05 | 0.01534432 |
|---|
| regulation of cardiac muscle cell proliferation | GO:0060043 |  | 4.71E-05 | 0.01416469 |
|---|
| regulation of phospholipase A2 activity | GO:0032429 |  | 4.71E-05 | 0.01259083 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 4.71E-05 | 0.01133175 |
|---|
| negative regulation of T cell mediated immunity | GO:0002710 |  | 4.71E-05 | 0.01030159 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of axonogenesis | GO:0050772 |  | 6.081E-06 | 0.01398732 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.604E-05 | 0.01844678 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.884E-05 | 0.0144414 |
|---|
| positive regulation of cell projection organization | GO:0031346 |  | 3.32E-05 | 0.01908854 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 3.32E-05 | 0.01527083 |
|---|
| regulation of cell morphogenesis | GO:0022604 |  | 4.686E-05 | 0.01796274 |
|---|
| cholesterol homeostasis | GO:0042632 |  | 5.334E-05 | 0.0175253 |
|---|
| regulation of axonogenesis | GO:0050770 |  | 5.938E-05 | 0.01707253 |
|---|
| cholesterol transport | GO:0030301 |  | 6.586E-05 | 0.01683071 |
|---|
| positive regulation of neurogenesis | GO:0050769 |  | 6.586E-05 | 0.01514764 |
|---|
| regulation of cardiac muscle cell proliferation | GO:0060043 |  | 6.739E-05 | 0.0140916 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of axonogenesis | GO:0050772 |  | 6.081E-06 | 0.01398732 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.604E-05 | 0.01844678 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.884E-05 | 0.0144414 |
|---|
| positive regulation of cell projection organization | GO:0031346 |  | 3.32E-05 | 0.01908854 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 3.32E-05 | 0.01527083 |
|---|
| regulation of cell morphogenesis | GO:0022604 |  | 4.686E-05 | 0.01796274 |
|---|
| cholesterol homeostasis | GO:0042632 |  | 5.334E-05 | 0.0175253 |
|---|
| regulation of axonogenesis | GO:0050770 |  | 5.938E-05 | 0.01707253 |
|---|
| cholesterol transport | GO:0030301 |  | 6.586E-05 | 0.01683071 |
|---|
| positive regulation of neurogenesis | GO:0050769 |  | 6.586E-05 | 0.01514764 |
|---|
| regulation of cardiac muscle cell proliferation | GO:0060043 |  | 6.739E-05 | 0.0140916 |
|---|


