Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 793
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3334 | 1.501e-03 | 2.600e-05 | 1.620e-01 | 6.324e-09 |
|---|
| IPC | 0.3393 | 3.514e-01 | 1.807e-01 | 9.362e-01 | 5.943e-02 |
|---|
| Loi | 0.4484 | 1.940e-04 | 0.000e+00 | 6.894e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5734 | 4.000e-06 | 0.000e+00 | 1.765e-01 | 0.000e+00 |
|---|
| Wang | 0.2396 | 1.508e-02 | 1.011e-01 | 3.120e-01 | 4.757e-04 |
|---|
Expression data for subnetwork 793 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
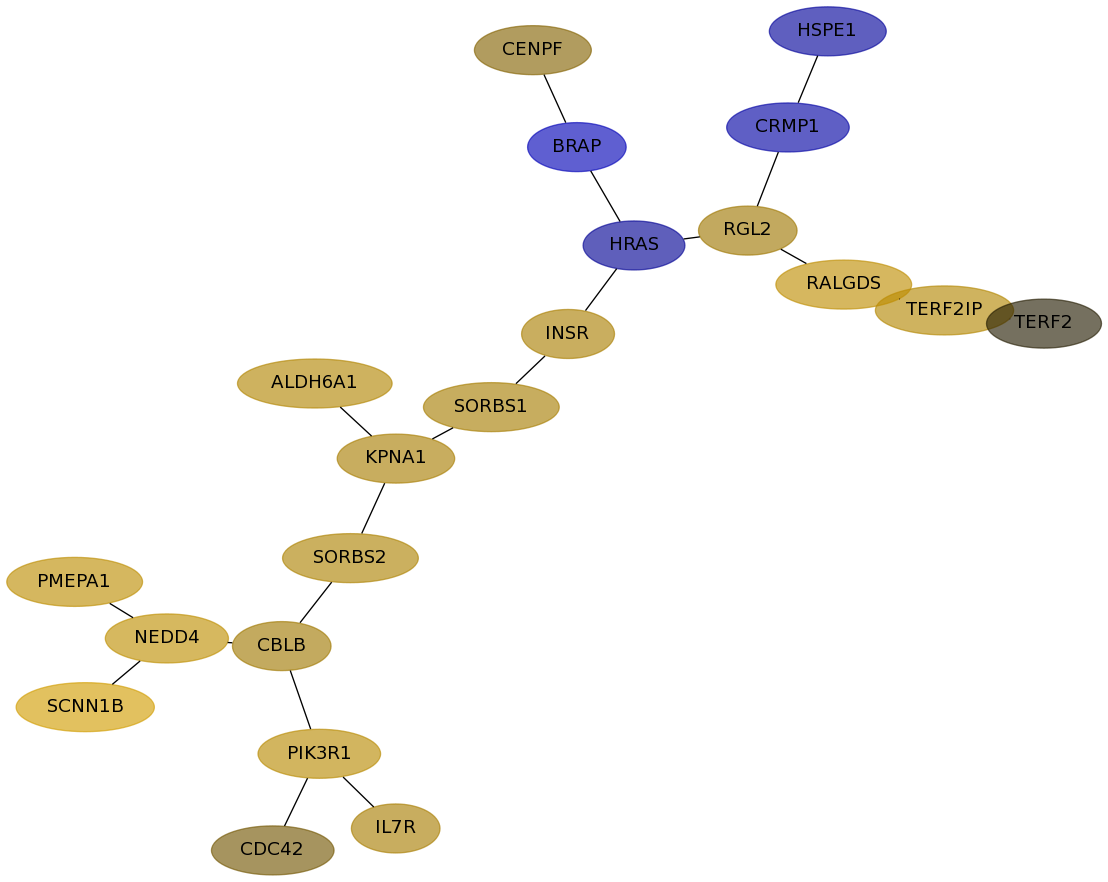
Score for each gene in subnetwork 793 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| SORBS1 |   | 17 | 30 | 182 | 178 | 0.124 | -0.043 | 0.015 | 0.314 | 0.200 |
|---|
| ALDH6A1 |   | 1 | 652 | 1451 | 1451 | 0.161 | -0.010 | 0.124 | 0.000 | 0.094 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| CENPF |   | 1 | 652 | 1451 | 1451 | 0.055 | 0.251 | -0.137 | 0.305 | 0.130 |
|---|
| CBLB |   | 3 | 265 | 182 | 189 | 0.109 | -0.168 | 0.266 | -0.248 | 0.223 |
|---|
| RALGDS |   | 5 | 148 | 647 | 621 | 0.212 | -0.011 | 0.199 | 0.033 | -0.009 |
|---|
| BRAP |   | 1 | 652 | 1451 | 1451 | -0.177 | 0.096 | -0.077 | -0.063 | -0.158 |
|---|
| SORBS2 |   | 3 | 265 | 778 | 770 | 0.147 | -0.075 | 0.077 | 0.084 | -0.114 |
|---|
| NEDD4 |   | 2 | 391 | 1385 | 1385 | 0.222 | -0.099 | 0.196 | -0.005 | 0.146 |
|---|
| HSPE1 |   | 1 | 652 | 1451 | 1451 | -0.092 | -0.067 | 0.063 | -0.146 | -0.220 |
|---|
| TERF2IP |   | 1 | 652 | 1451 | 1451 | 0.168 | -0.201 | 0.128 | -0.042 | 0.087 |
|---|
| KPNA1 |   | 3 | 265 | 1198 | 1173 | 0.130 | 0.007 | 0.152 | -0.083 | 0.086 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| PMEPA1 |   | 4 | 198 | 937 | 910 | 0.208 | -0.085 | 0.137 | 0.167 | 0.102 |
|---|
| IL7R |   | 14 | 42 | 339 | 334 | 0.130 | -0.151 | -0.112 | -0.130 | -0.160 |
|---|
| RGL2 |   | 3 | 265 | 647 | 632 | 0.105 | -0.107 | 0.227 | -0.146 | -0.002 |
|---|
| SCNN1B |   | 2 | 391 | 1385 | 1385 | 0.336 | -0.246 | 0.073 | -0.147 | -0.039 |
|---|
| CRMP1 |   | 3 | 265 | 647 | 632 | -0.114 | -0.068 | 0.174 | 0.049 | 0.151 |
|---|
| HRAS |   | 3 | 265 | 1094 | 1070 | -0.081 | 0.037 | 0.090 | 0.098 | -0.005 |
|---|
| TERF2 |   | 1 | 652 | 1451 | 1451 | 0.005 | -0.252 | -0.034 | -0.033 | 0.262 |
|---|
GO Enrichment output for subnetwork 793 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 3.714E-08 | 8.542E-05 |
|---|
| nuclear migration | GO:0007097 |  | 1.106E-07 | 1.272E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.106E-07 | 8.481E-05 |
|---|
| macrophage differentiation | GO:0030225 |  | 2.209E-07 | 1.27E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.415E-07 | 1.111E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 3.859E-07 | 1.479E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 3.859E-07 | 1.268E-04 |
|---|
| negative regulation of telomere maintenance | GO:0032205 |  | 3.859E-07 | 1.11E-04 |
|---|
| regulation of telomere maintenance | GO:0032204 |  | 6.165E-07 | 1.576E-04 |
|---|
| telomere maintenance via telomerase | GO:0007004 |  | 9.233E-07 | 2.124E-04 |
|---|
| nucleus localization | GO:0051647 |  | 9.233E-07 | 1.931E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.919E-08 | 4.688E-05 |
|---|
| nuclear migration | GO:0007097 |  | 1.007E-07 | 1.23E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.007E-07 | 8.2E-05 |
|---|
| interaction with host | GO:0051701 |  | 1.033E-07 | 6.306E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.429E-07 | 6.981E-05 |
|---|
| negative regulation of telomere maintenance | GO:0032205 |  | 1.76E-07 | 7.166E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.813E-07 | 9.816E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.813E-07 | 8.589E-05 |
|---|
| symbiosis. encompassing mutualism through parasitism | GO:0044403 |  | 3.741E-07 | 1.016E-04 |
|---|
| regulation of telomere maintenance | GO:0032204 |  | 4.214E-07 | 1.029E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 4.214E-07 | 9.358E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 3.033E-08 | 7.297E-05 |
|---|
| nuclear migration | GO:0007097 |  | 1.28E-07 | 1.54E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.28E-07 | 1.027E-04 |
|---|
| interaction with host | GO:0051701 |  | 1.415E-07 | 8.514E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.67E-07 | 8.038E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.238E-07 | 8.972E-05 |
|---|
| macrophage differentiation | GO:0030225 |  | 2.238E-07 | 7.691E-05 |
|---|
| negative regulation of telomere maintenance | GO:0032205 |  | 2.238E-07 | 6.729E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 3.575E-07 | 9.558E-05 |
|---|
| symbiosis. encompassing mutualism through parasitism | GO:0044403 |  | 5.124E-07 | 1.233E-04 |
|---|
| regulation of telomere maintenance | GO:0032204 |  | 5.356E-07 | 1.171E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 3.714E-08 | 8.542E-05 |
|---|
| nuclear migration | GO:0007097 |  | 1.106E-07 | 1.272E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.106E-07 | 8.481E-05 |
|---|
| macrophage differentiation | GO:0030225 |  | 2.209E-07 | 1.27E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.415E-07 | 1.111E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 3.859E-07 | 1.479E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 3.859E-07 | 1.268E-04 |
|---|
| negative regulation of telomere maintenance | GO:0032205 |  | 3.859E-07 | 1.11E-04 |
|---|
| regulation of telomere maintenance | GO:0032204 |  | 6.165E-07 | 1.576E-04 |
|---|
| telomere maintenance via telomerase | GO:0007004 |  | 9.233E-07 | 2.124E-04 |
|---|
| nucleus localization | GO:0051647 |  | 9.233E-07 | 1.931E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 3.714E-08 | 8.542E-05 |
|---|
| nuclear migration | GO:0007097 |  | 1.106E-07 | 1.272E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.106E-07 | 8.481E-05 |
|---|
| macrophage differentiation | GO:0030225 |  | 2.209E-07 | 1.27E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.415E-07 | 1.111E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 3.859E-07 | 1.479E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 3.859E-07 | 1.268E-04 |
|---|
| negative regulation of telomere maintenance | GO:0032205 |  | 3.859E-07 | 1.11E-04 |
|---|
| regulation of telomere maintenance | GO:0032204 |  | 6.165E-07 | 1.576E-04 |
|---|
| telomere maintenance via telomerase | GO:0007004 |  | 9.233E-07 | 2.124E-04 |
|---|
| nucleus localization | GO:0051647 |  | 9.233E-07 | 1.931E-04 |
|---|


