Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 787
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3326 | 1.557e-03 | 2.800e-05 | 1.646e-01 | 7.177e-09 |
|---|
| IPC | 0.3416 | 3.474e-01 | 1.772e-01 | 9.354e-01 | 5.760e-02 |
|---|
| Loi | 0.4478 | 1.980e-04 | 0.000e+00 | 6.930e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5755 | 3.000e-06 | 0.000e+00 | 1.709e-01 | 0.000e+00 |
|---|
| Wang | 0.2400 | 1.485e-02 | 1.002e-01 | 3.099e-01 | 4.608e-04 |
|---|
Expression data for subnetwork 787 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
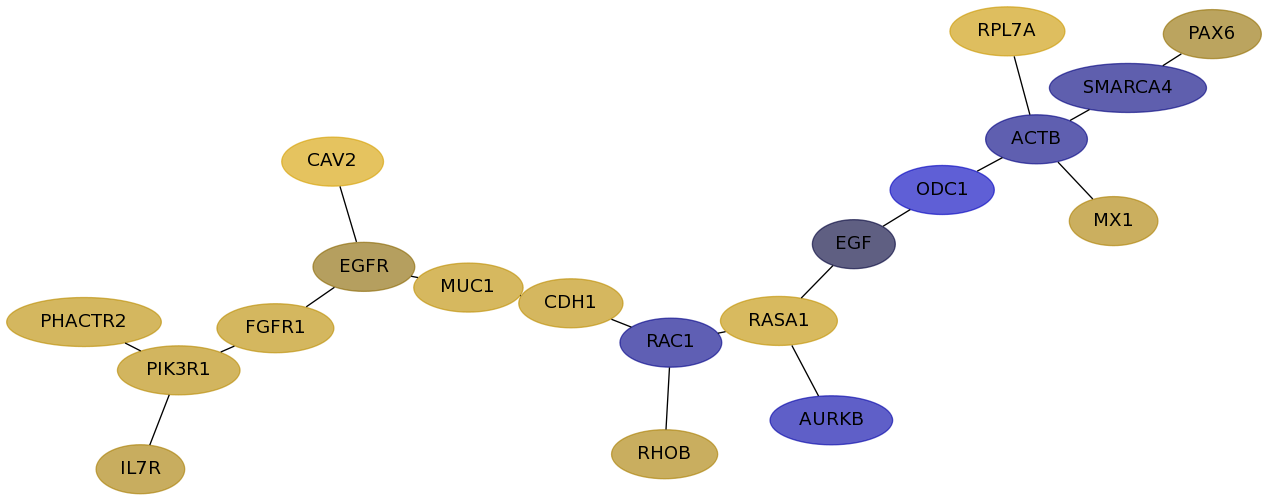
Score for each gene in subnetwork 787 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| MX1 |   | 3 | 265 | 398 | 404 | 0.143 | 0.092 | -0.016 | -0.165 | -0.267 |
|---|
| PAX6 |   | 11 | 59 | 283 | 274 | 0.081 | 0.259 | -0.003 | 0.112 | 0.168 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| RAC1 |   | 1 | 652 | 1448 | 1448 | -0.061 | 0.062 | 0.158 | 0.003 | -0.031 |
|---|
| EGF |   | 11 | 59 | 398 | 383 | -0.009 | -0.212 | 0.182 | -0.276 | 0.005 |
|---|
| SMARCA4 |   | 4 | 198 | 283 | 289 | -0.050 | 0.080 | 0.202 | -0.071 | -0.116 |
|---|
| PHACTR2 |   | 5 | 148 | 271 | 267 | 0.203 | -0.057 | -0.051 | 0.143 | 0.164 |
|---|
| AURKB |   | 3 | 265 | 1236 | 1219 | -0.132 | 0.179 | -0.189 | 0.086 | -0.028 |
|---|
| ODC1 |   | 5 | 148 | 398 | 387 | -0.220 | -0.006 | -0.099 | 0.209 | -0.107 |
|---|
| RHOB |   | 1 | 652 | 1448 | 1448 | 0.135 | -0.018 | 0.109 | -0.008 | 0.015 |
|---|
| RASA1 |   | 9 | 77 | 492 | 494 | 0.237 | -0.079 | 0.146 | 0.127 | 0.149 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| RPL7A |   | 1 | 652 | 1448 | 1448 | 0.292 | 0.005 | 0.111 | 0.214 | -0.044 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| IL7R |   | 14 | 42 | 339 | 334 | 0.130 | -0.151 | -0.112 | -0.130 | -0.160 |
|---|
| ACTB |   | 7 | 100 | 398 | 385 | -0.052 | -0.039 | 0.283 | -0.061 | 0.003 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
GO Enrichment output for subnetwork 787 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| localization within membrane | GO:0051668 |  | 4.356E-09 | 1.002E-05 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 1.482E-07 | 1.705E-04 |
|---|
| negative regulation of endocytosis | GO:0045806 |  | 2.959E-07 | 2.269E-04 |
|---|
| positive regulation of small GTPase mediated signal transduction | GO:0051057 |  | 8.257E-07 | 4.748E-04 |
|---|
| ruffle organization | GO:0031529 |  | 8.257E-07 | 3.798E-04 |
|---|
| lamellipodium assembly | GO:0030032 |  | 8.257E-07 | 3.165E-04 |
|---|
| actin filament polymerization | GO:0030041 |  | 2.42E-06 | 7.952E-04 |
|---|
| actin polymerization or depolymerization | GO:0008154 |  | 9.869E-06 | 2.837E-03 |
|---|
| regulation of receptor-mediated endocytosis | GO:0048259 |  | 1.646E-05 | 4.206E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.646E-05 | 3.785E-03 |
|---|
| vasculogenesis | GO:0001570 |  | 2.216E-05 | 4.632E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| localization within membrane | GO:0051668 |  | 9.878E-12 | 2.413E-08 |
|---|
| negative regulation of receptor-mediated endocytosis | GO:0048261 |  | 3.919E-11 | 4.787E-08 |
|---|
| regulation of hydrogen peroxide metabolic process | GO:0010310 |  | 1.174E-10 | 9.562E-08 |
|---|
| regulation of oxygen and reactive oxygen species metabolic process | GO:0080010 |  | 2.736E-10 | 1.671E-07 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 5.466E-10 | 2.67E-07 |
|---|
| negative regulation of endocytosis | GO:0045806 |  | 9.825E-10 | 4.001E-07 |
|---|
| ruffle organization | GO:0031529 |  | 1.635E-09 | 5.708E-07 |
|---|
| lamellipodium assembly | GO:0030032 |  | 3.845E-09 | 1.174E-06 |
|---|
| positive regulation of small GTPase mediated signal transduction | GO:0051057 |  | 7.756E-09 | 2.105E-06 |
|---|
| actin filament polymerization | GO:0030041 |  | 1.056E-08 | 2.58E-06 |
|---|
| actin polymerization or depolymerization | GO:0008154 |  | 5.61E-08 | 1.246E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| localization within membrane | GO:0051668 |  | 2.4E-09 | 5.774E-06 |
|---|
| regulation of hydrogen peroxide metabolic process | GO:0010310 |  | 6.41E-08 | 7.711E-05 |
|---|
| regulation of oxygen and reactive oxygen species metabolic process | GO:0080010 |  | 1.28E-07 | 1.027E-04 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 2.238E-07 | 1.346E-04 |
|---|
| negative regulation of endocytosis | GO:0045806 |  | 3.575E-07 | 1.72E-04 |
|---|
| ruffle organization | GO:0031529 |  | 5.356E-07 | 2.148E-04 |
|---|
| positive regulation of small GTPase mediated signal transduction | GO:0051057 |  | 1.049E-06 | 3.606E-04 |
|---|
| lamellipodium assembly | GO:0030032 |  | 1.049E-06 | 3.155E-04 |
|---|
| actin filament polymerization | GO:0030041 |  | 1.814E-06 | 4.849E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 7.162E-06 | 1.723E-03 |
|---|
| actin polymerization or depolymerization | GO:0008154 |  | 7.162E-06 | 1.566E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| localization within membrane | GO:0051668 |  | 4.356E-09 | 1.002E-05 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 1.482E-07 | 1.705E-04 |
|---|
| negative regulation of endocytosis | GO:0045806 |  | 2.959E-07 | 2.269E-04 |
|---|
| positive regulation of small GTPase mediated signal transduction | GO:0051057 |  | 8.257E-07 | 4.748E-04 |
|---|
| ruffle organization | GO:0031529 |  | 8.257E-07 | 3.798E-04 |
|---|
| lamellipodium assembly | GO:0030032 |  | 8.257E-07 | 3.165E-04 |
|---|
| actin filament polymerization | GO:0030041 |  | 2.42E-06 | 7.952E-04 |
|---|
| actin polymerization or depolymerization | GO:0008154 |  | 9.869E-06 | 2.837E-03 |
|---|
| regulation of receptor-mediated endocytosis | GO:0048259 |  | 1.646E-05 | 4.206E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.646E-05 | 3.785E-03 |
|---|
| vasculogenesis | GO:0001570 |  | 2.216E-05 | 4.632E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| localization within membrane | GO:0051668 |  | 4.356E-09 | 1.002E-05 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 1.482E-07 | 1.705E-04 |
|---|
| negative regulation of endocytosis | GO:0045806 |  | 2.959E-07 | 2.269E-04 |
|---|
| positive regulation of small GTPase mediated signal transduction | GO:0051057 |  | 8.257E-07 | 4.748E-04 |
|---|
| ruffle organization | GO:0031529 |  | 8.257E-07 | 3.798E-04 |
|---|
| lamellipodium assembly | GO:0030032 |  | 8.257E-07 | 3.165E-04 |
|---|
| actin filament polymerization | GO:0030041 |  | 2.42E-06 | 7.952E-04 |
|---|
| actin polymerization or depolymerization | GO:0008154 |  | 9.869E-06 | 2.837E-03 |
|---|
| regulation of receptor-mediated endocytosis | GO:0048259 |  | 1.646E-05 | 4.206E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.646E-05 | 3.785E-03 |
|---|
| vasculogenesis | GO:0001570 |  | 2.216E-05 | 4.632E-03 |
|---|


