Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 753
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3270 | 1.994e-03 | 4.200e-05 | 1.830e-01 | 1.533e-08 |
|---|
| IPC | 0.3518 | 3.308e-01 | 1.630e-01 | 9.319e-01 | 5.023e-02 |
|---|
| Loi | 0.4468 | 2.080e-04 | 0.000e+00 | 6.994e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5827 | 2.000e-06 | 0.000e+00 | 1.523e-01 | 0.000e+00 |
|---|
| Wang | 0.2388 | 1.549e-02 | 1.028e-01 | 3.156e-01 | 5.026e-04 |
|---|
Expression data for subnetwork 753 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
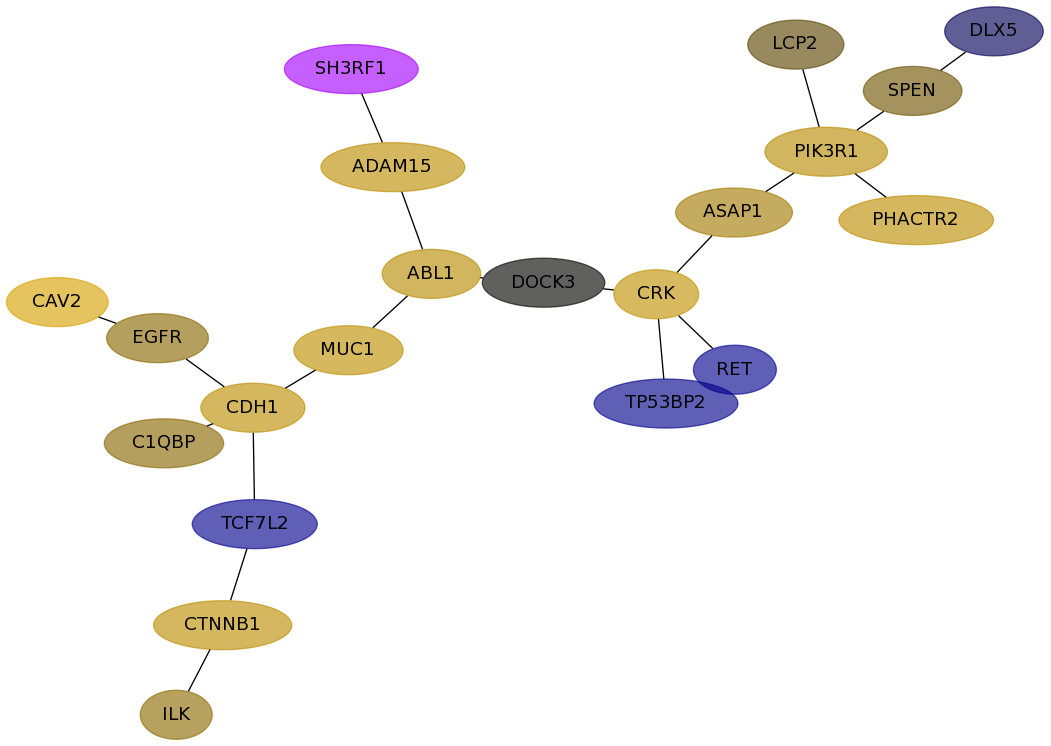
Score for each gene in subnetwork 753 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| RET |   | 4 | 198 | 57 | 68 | -0.069 | 0.098 | 0.063 | -0.179 | -0.159 |
|---|
| PHACTR2 |   | 5 | 148 | 271 | 267 | 0.203 | -0.057 | -0.051 | 0.143 | 0.164 |
|---|
| ILK |   | 6 | 120 | 33 | 40 | 0.069 | -0.020 | 0.195 | 0.076 | 0.086 |
|---|
| LCP2 |   | 1 | 652 | 1443 | 1443 | 0.021 | 0.087 | -0.303 | -0.077 | -0.047 |
|---|
| DOCK3 |   | 1 | 652 | 1443 | 1443 | 0.000 | -0.174 | 0.073 | -0.106 | -0.077 |
|---|
| C1QBP |   | 4 | 198 | 552 | 547 | 0.064 | 0.134 | -0.072 | 0.080 | -0.006 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| TCF7L2 |   | 17 | 30 | 271 | 264 | -0.071 | -0.021 | 0.143 | 0.280 | 0.101 |
|---|
| ABL1 |   | 8 | 84 | 326 | 316 | 0.193 | -0.152 | 0.200 | 0.108 | -0.059 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CRK |   | 27 | 17 | 206 | 193 | 0.232 | -0.116 | 0.106 | 0.090 | 0.017 |
|---|
| SH3RF1 |   | 14 | 42 | 117 | 120 | undef | 0.144 | 0.238 | undef | undef |
|---|
| ASAP1 |   | 6 | 120 | 129 | 128 | 0.114 | 0.036 | 0.070 | 0.035 | -0.056 |
|---|
| SPEN |   | 3 | 265 | 129 | 139 | 0.034 | -0.147 | 0.207 | -0.086 | 0.238 |
|---|
| ADAM15 |   | 13 | 47 | 492 | 474 | 0.205 | -0.048 | -0.078 | -0.208 | -0.048 |
|---|
| DLX5 |   | 2 | 391 | 1008 | 999 | -0.019 | -0.026 | 0.044 | -0.151 | 0.140 |
|---|
| TP53BP2 |   | 5 | 148 | 1 | 22 | -0.067 | 0.245 | -0.012 | 0.129 | 0.017 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
GO Enrichment output for subnetwork 753 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 6.651E-07 | 1.53E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.13E-05 | 0.01299862 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.328E-05 | 0.01017839 |
|---|
| ureteric bud development | GO:0001657 |  | 1.788E-05 | 0.01027875 |
|---|
| regulation of oxidoreductase activity | GO:0051341 |  | 2.342E-05 | 0.0107722 |
|---|
| metanephros development | GO:0001656 |  | 5.142E-05 | 0.01971012 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 5.343E-05 | 0.01755404 |
|---|
| enteric nervous system development | GO:0048484 |  | 5.343E-05 | 0.01535979 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 5.343E-05 | 0.01365314 |
|---|
| protein homooligomerization | GO:0051260 |  | 7.438E-05 | 0.01710767 |
|---|
| positive regulation of insulin secretion | GO:0032024 |  | 8.002E-05 | 0.01673122 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.912E-11 | 4.671E-08 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 1.946E-07 | 2.377E-04 |
|---|
| positive regulation of insulin secretion | GO:0032024 |  | 5.431E-07 | 4.423E-04 |
|---|
| positive regulation of peptide secretion | GO:0002793 |  | 1.593E-06 | 9.728E-04 |
|---|
| mesenchymal cell development | GO:0014031 |  | 1.939E-06 | 9.476E-04 |
|---|
| regulation of hormone metabolic process | GO:0032350 |  | 2.12E-06 | 8.634E-04 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 4.365E-06 | 1.523E-03 |
|---|
| myoblast differentiation | GO:0045445 |  | 6.503E-06 | 1.986E-03 |
|---|
| positive regulation of hormone secretion | GO:0046887 |  | 9.238E-06 | 2.508E-03 |
|---|
| pancreas development | GO:0031016 |  | 1.085E-05 | 2.651E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.264E-05 | 2.807E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.36E-11 | 3.272E-08 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 2.29E-07 | 2.755E-04 |
|---|
| positive regulation of insulin secretion | GO:0032024 |  | 6.392E-07 | 5.127E-04 |
|---|
| positive regulation of peptide secretion | GO:0002793 |  | 1.874E-06 | 1.127E-03 |
|---|
| mesenchymal cell development | GO:0014031 |  | 2.166E-06 | 1.042E-03 |
|---|
| regulation of hormone metabolic process | GO:0032350 |  | 2.495E-06 | 1E-03 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 5.134E-06 | 1.765E-03 |
|---|
| myoblast differentiation | GO:0045445 |  | 7.648E-06 | 2.3E-03 |
|---|
| positive regulation of hormone secretion | GO:0046887 |  | 1.086E-05 | 2.904E-03 |
|---|
| pancreas development | GO:0031016 |  | 1.276E-05 | 3.07E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.276E-05 | 2.791E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 6.651E-07 | 1.53E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.13E-05 | 0.01299862 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.328E-05 | 0.01017839 |
|---|
| ureteric bud development | GO:0001657 |  | 1.788E-05 | 0.01027875 |
|---|
| regulation of oxidoreductase activity | GO:0051341 |  | 2.342E-05 | 0.0107722 |
|---|
| metanephros development | GO:0001656 |  | 5.142E-05 | 0.01971012 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 5.343E-05 | 0.01755404 |
|---|
| enteric nervous system development | GO:0048484 |  | 5.343E-05 | 0.01535979 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 5.343E-05 | 0.01365314 |
|---|
| protein homooligomerization | GO:0051260 |  | 7.438E-05 | 0.01710767 |
|---|
| positive regulation of insulin secretion | GO:0032024 |  | 8.002E-05 | 0.01673122 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 6.651E-07 | 1.53E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.13E-05 | 0.01299862 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.328E-05 | 0.01017839 |
|---|
| ureteric bud development | GO:0001657 |  | 1.788E-05 | 0.01027875 |
|---|
| regulation of oxidoreductase activity | GO:0051341 |  | 2.342E-05 | 0.0107722 |
|---|
| metanephros development | GO:0001656 |  | 5.142E-05 | 0.01971012 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 5.343E-05 | 0.01755404 |
|---|
| enteric nervous system development | GO:0048484 |  | 5.343E-05 | 0.01535979 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 5.343E-05 | 0.01365314 |
|---|
| protein homooligomerization | GO:0051260 |  | 7.438E-05 | 0.01710767 |
|---|
| positive regulation of insulin secretion | GO:0032024 |  | 8.002E-05 | 0.01673122 |
|---|


