Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 704
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3136 | 3.533e-03 | 1.070e-04 | 2.324e-01 | 8.784e-08 |
|---|
| IPC | 0.3728 | 2.964e-01 | 1.352e-01 | 9.245e-01 | 3.703e-02 |
|---|
| Loi | 0.4446 | 2.280e-04 | 0.000e+00 | 7.131e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5990 | 1.000e-06 | 0.000e+00 | 1.148e-01 | 0.000e+00 |
|---|
| Wang | 0.2393 | 1.522e-02 | 1.017e-01 | 3.133e-01 | 4.849e-04 |
|---|
Expression data for subnetwork 704 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
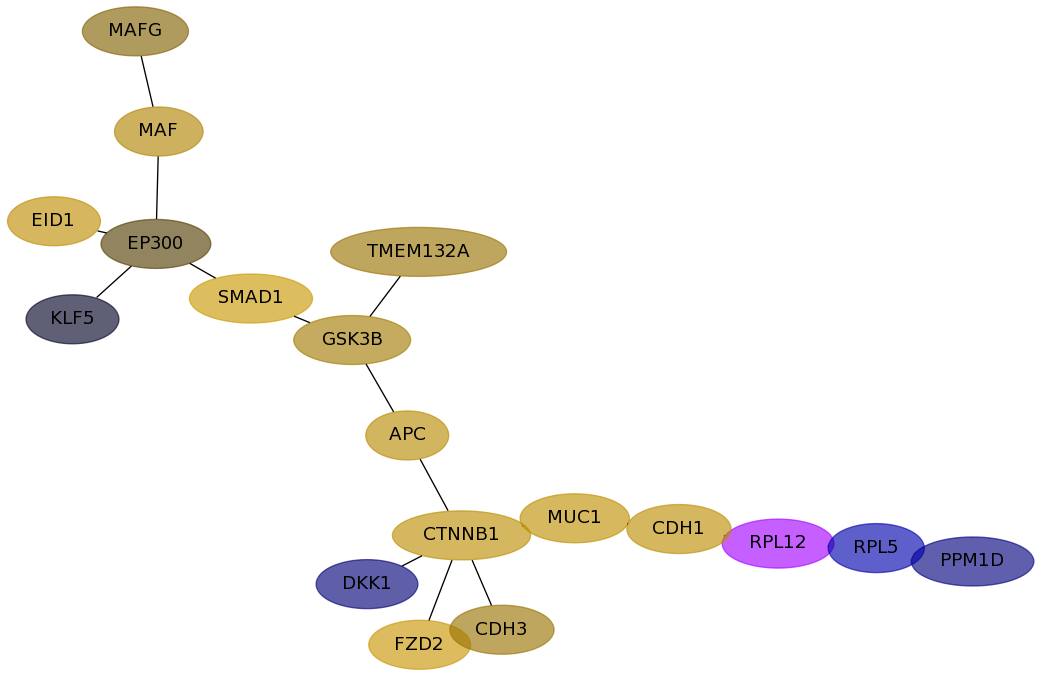
Score for each gene in subnetwork 704 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| RPL12 |   | 1 | 652 | 1417 | 1417 | undef | -0.145 | -0.025 | undef | undef |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| EID1 |   | 4 | 198 | 552 | 547 | 0.212 | -0.069 | 0.051 | -0.010 | -0.037 |
|---|
| PPM1D |   | 2 | 391 | 889 | 891 | -0.050 | 0.105 | 0.121 | 0.125 | 0.166 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| CDH3 |   | 10 | 67 | 1 | 16 | 0.088 | 0.028 | 0.169 | -0.055 | -0.027 |
|---|
| KLF5 |   | 25 | 19 | 57 | 50 | -0.005 | 0.077 | 0.117 | -0.007 | -0.006 |
|---|
| APC |   | 1 | 652 | 1417 | 1417 | 0.189 | -0.006 | -0.004 | 0.067 | 0.139 |
|---|
| SMAD1 |   | 24 | 21 | 33 | 30 | 0.287 | 0.049 | 0.159 | -0.062 | 0.048 |
|---|
| TMEM132A |   | 7 | 100 | 33 | 37 | 0.088 | 0.019 | 0.127 | -0.141 | -0.092 |
|---|
| FZD2 |   | 8 | 84 | 552 | 540 | 0.273 | -0.067 | -0.081 | -0.067 | 0.081 |
|---|
| MAF |   | 2 | 391 | 552 | 554 | 0.155 | 0.024 | 0.085 | -0.094 | 0.122 |
|---|
| RPL5 |   | 4 | 198 | 889 | 873 | -0.145 | -0.000 | 0.128 | 0.220 | -0.110 |
|---|
| MAFG |   | 1 | 652 | 1417 | 1417 | 0.052 | -0.138 | -0.087 | 0.180 | 0.074 |
|---|
| DKK1 |   | 4 | 198 | 271 | 273 | -0.041 | 0.134 | 0.091 | -0.124 | 0.129 |
|---|
GO Enrichment output for subnetwork 704 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 8.299E-10 | 1.909E-06 |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 5.836E-09 | 6.711E-06 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 6.749E-07 | 5.174E-04 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 4.486E-06 | 2.579E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 6.376E-06 | 2.933E-03 |
|---|
| negative regulation of Wnt receptor signaling pathway | GO:0030178 |  | 8.728E-06 | 3.346E-03 |
|---|
| rRNA metabolic process | GO:0016072 |  | 2.159E-05 | 7.095E-03 |
|---|
| regulation of protein localization | GO:0032880 |  | 2.159E-05 | 6.209E-03 |
|---|
| mitotic cell cycle spindle assembly checkpoint | GO:0007094 |  | 3.658E-05 | 9.348E-03 |
|---|
| microvillus organization | GO:0032528 |  | 3.658E-05 | 8.413E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.658E-05 | 7.649E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.332E-12 | 3.255E-09 |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 1.74E-09 | 2.125E-06 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 9.129E-07 | 7.434E-04 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 2.277E-06 | 1.391E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 3.07E-06 | 1.5E-03 |
|---|
| negative regulation of Wnt receptor signaling pathway | GO:0030178 |  | 3.528E-06 | 1.436E-03 |
|---|
| regulation of protein localization | GO:0032880 |  | 7.788E-06 | 2.718E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.528E-05 | 4.667E-03 |
|---|
| microvillus organization | GO:0032528 |  | 1.634E-05 | 4.437E-03 |
|---|
| regulation of chondrocyte differentiation | GO:0032330 |  | 1.634E-05 | 3.993E-03 |
|---|
| somatic stem cell maintenance | GO:0035019 |  | 1.634E-05 | 3.63E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 9.104E-13 | 2.19E-09 |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 2.094E-09 | 2.519E-06 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 1.053E-06 | 8.448E-04 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 2.627E-06 | 1.58E-03 |
|---|
| negative regulation of Wnt receptor signaling pathway | GO:0030178 |  | 3.062E-06 | 1.473E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 3.542E-06 | 1.42E-03 |
|---|
| regulation of protein localization | GO:0032880 |  | 9.344E-06 | 3.212E-03 |
|---|
| regulation of Wnt receptor signaling pathway | GO:0030111 |  | 1.621E-05 | 4.874E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.621E-05 | 4.332E-03 |
|---|
| microvillus organization | GO:0032528 |  | 1.803E-05 | 4.339E-03 |
|---|
| somatic stem cell maintenance | GO:0035019 |  | 1.803E-05 | 3.945E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 8.299E-10 | 1.909E-06 |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 5.836E-09 | 6.711E-06 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 6.749E-07 | 5.174E-04 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 4.486E-06 | 2.579E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 6.376E-06 | 2.933E-03 |
|---|
| negative regulation of Wnt receptor signaling pathway | GO:0030178 |  | 8.728E-06 | 3.346E-03 |
|---|
| rRNA metabolic process | GO:0016072 |  | 2.159E-05 | 7.095E-03 |
|---|
| regulation of protein localization | GO:0032880 |  | 2.159E-05 | 6.209E-03 |
|---|
| mitotic cell cycle spindle assembly checkpoint | GO:0007094 |  | 3.658E-05 | 9.348E-03 |
|---|
| microvillus organization | GO:0032528 |  | 3.658E-05 | 8.413E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.658E-05 | 7.649E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 8.299E-10 | 1.909E-06 |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 5.836E-09 | 6.711E-06 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 6.749E-07 | 5.174E-04 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 4.486E-06 | 2.579E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 6.376E-06 | 2.933E-03 |
|---|
| negative regulation of Wnt receptor signaling pathway | GO:0030178 |  | 8.728E-06 | 3.346E-03 |
|---|
| rRNA metabolic process | GO:0016072 |  | 2.159E-05 | 7.095E-03 |
|---|
| regulation of protein localization | GO:0032880 |  | 2.159E-05 | 6.209E-03 |
|---|
| mitotic cell cycle spindle assembly checkpoint | GO:0007094 |  | 3.658E-05 | 9.348E-03 |
|---|
| microvillus organization | GO:0032528 |  | 3.658E-05 | 8.413E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.658E-05 | 7.649E-03 |
|---|


