Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 678
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3030 | 5.410e-03 | 2.170e-04 | 2.757e-01 | 3.237e-07 |
|---|
| IPC | 0.3932 | 2.636e-01 | 1.108e-01 | 9.173e-01 | 2.679e-02 |
|---|
| Loi | 0.4420 | 2.550e-04 | 0.000e+00 | 7.293e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6102 | 1.000e-06 | 0.000e+00 | 9.330e-02 | 0.000e+00 |
|---|
| Wang | 0.2398 | 1.494e-02 | 1.006e-01 | 3.107e-01 | 4.670e-04 |
|---|
Expression data for subnetwork 678 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
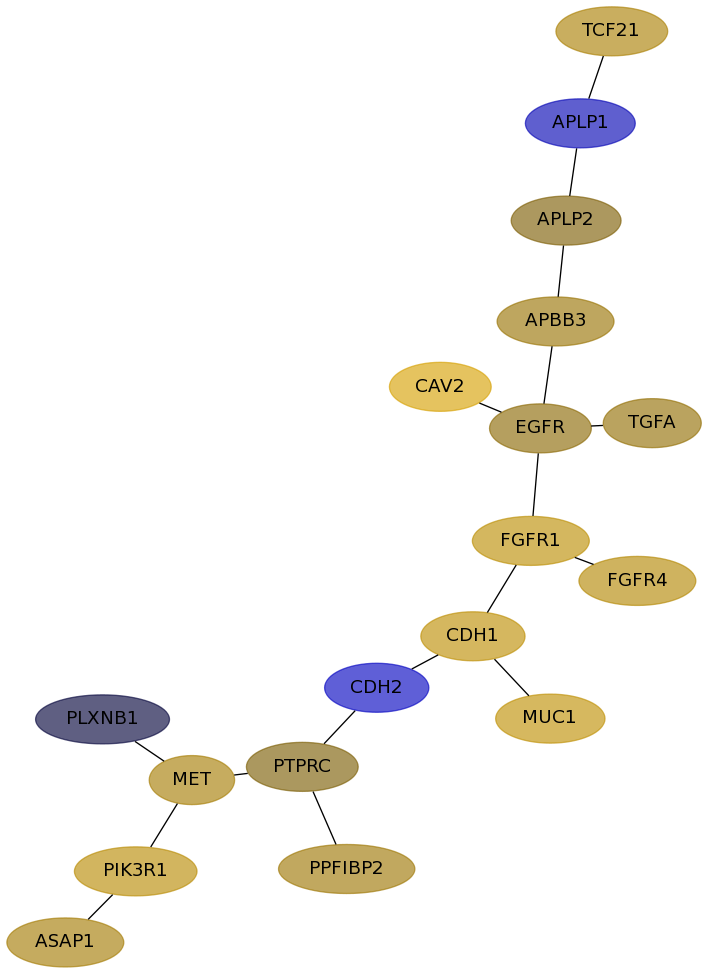
Score for each gene in subnetwork 678 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| TGFA |   | 8 | 84 | 439 | 429 | 0.077 | -0.057 | -0.027 | -0.158 | 0.020 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| PPFIBP2 |   | 7 | 100 | 606 | 576 | 0.098 | 0.188 | 0.320 | -0.112 | 0.013 |
|---|
| TCF21 |   | 1 | 652 | 1232 | 1235 | 0.133 | 0.135 | 0.142 | -0.088 | -0.092 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| PLXNB1 |   | 2 | 391 | 1232 | 1220 | -0.009 | -0.117 | 0.312 | 0.006 | -0.146 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| PTPRC |   | 5 | 148 | 715 | 697 | 0.043 | -0.031 | -0.204 | -0.050 | -0.068 |
|---|
| APBB3 |   | 1 | 652 | 1232 | 1235 | 0.090 | 0.039 | 0.161 | -0.024 | 0.035 |
|---|
| APLP2 |   | 1 | 652 | 1232 | 1235 | 0.045 | -0.249 | 0.198 | -0.163 | -0.104 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| APLP1 |   | 3 | 265 | 859 | 843 | -0.170 | 0.284 | 0.043 | -0.105 | -0.190 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| ASAP1 |   | 6 | 120 | 129 | 128 | 0.114 | 0.036 | 0.070 | 0.035 | -0.056 |
|---|
| MET |   | 5 | 148 | 536 | 536 | 0.119 | -0.219 | 0.086 | 0.183 | 0.170 |
|---|
| CDH2 |   | 7 | 100 | 33 | 37 | -0.228 | 0.169 | 0.109 | -0.016 | 0.155 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
GO Enrichment output for subnetwork 678 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.825E-05 | 0.0419786 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.143E-05 | 0.02464568 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 3.776E-05 | 0.02894696 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 7.343E-05 | 0.04222427 |
|---|
| negative regulation of T cell mediated immunity | GO:0002710 |  | 7.343E-05 | 0.03377942 |
|---|
| negative regulation of peptidyl-tyrosine phosphorylation | GO:0050732 |  | 7.343E-05 | 0.02814952 |
|---|
| positive regulation of T cell mediated cytotoxicity | GO:0001916 |  | 7.343E-05 | 0.02412816 |
|---|
| release of sequestered calcium ion into cytosol | GO:0051209 |  | 7.343E-05 | 0.02111214 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 7.343E-05 | 0.01876634 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 8.603E-05 | 0.01978746 |
|---|
| thymic T cell selection | GO:0045061 |  | 1.1E-04 | 0.02299104 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA integration | GO:0015074 |  | 6.633E-06 | 0.01620467 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.076E-05 | 0.01314765 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.631E-05 | 0.01327983 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.922E-05 | 0.01784874 |
|---|
| regulation of phospholipase A2 activity | GO:0032429 |  | 4.144E-05 | 0.02024661 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 4.144E-05 | 0.01687218 |
|---|
| negative regulation of T cell mediated immunity | GO:0002710 |  | 4.144E-05 | 0.01446187 |
|---|
| negative regulation of peptidyl-tyrosine phosphorylation | GO:0050732 |  | 4.144E-05 | 0.01265413 |
|---|
| positive regulation of T cell mediated cytotoxicity | GO:0001916 |  | 4.144E-05 | 0.01124812 |
|---|
| release of sequestered calcium ion into cytosol | GO:0051209 |  | 4.144E-05 | 0.01012331 |
|---|
| protein insertion into membrane | GO:0051205 |  | 4.144E-05 | 9.203E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA integration | GO:0015074 |  | 6.687E-06 | 0.0160882 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.116E-05 | 0.01342239 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.503E-05 | 0.01205015 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.233E-05 | 0.0134344 |
|---|
| regulation of phospholipase A2 activity | GO:0032429 |  | 4.71E-05 | 0.0226635 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 4.71E-05 | 0.01888625 |
|---|
| negative regulation of T cell mediated immunity | GO:0002710 |  | 4.71E-05 | 0.01618821 |
|---|
| negative regulation of peptidyl-tyrosine phosphorylation | GO:0050732 |  | 4.71E-05 | 0.01416469 |
|---|
| positive regulation of T cell mediated cytotoxicity | GO:0001916 |  | 4.71E-05 | 0.01259083 |
|---|
| release of sequestered calcium ion into cytosol | GO:0051209 |  | 4.71E-05 | 0.01133175 |
|---|
| regulation of ARF GTPase activity | GO:0032312 |  | 4.764E-05 | 0.01042017 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.825E-05 | 0.0419786 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.143E-05 | 0.02464568 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 3.776E-05 | 0.02894696 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 7.343E-05 | 0.04222427 |
|---|
| negative regulation of T cell mediated immunity | GO:0002710 |  | 7.343E-05 | 0.03377942 |
|---|
| negative regulation of peptidyl-tyrosine phosphorylation | GO:0050732 |  | 7.343E-05 | 0.02814952 |
|---|
| positive regulation of T cell mediated cytotoxicity | GO:0001916 |  | 7.343E-05 | 0.02412816 |
|---|
| release of sequestered calcium ion into cytosol | GO:0051209 |  | 7.343E-05 | 0.02111214 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 7.343E-05 | 0.01876634 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 8.603E-05 | 0.01978746 |
|---|
| thymic T cell selection | GO:0045061 |  | 1.1E-04 | 0.02299104 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.825E-05 | 0.0419786 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.143E-05 | 0.02464568 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 3.776E-05 | 0.02894696 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 7.343E-05 | 0.04222427 |
|---|
| negative regulation of T cell mediated immunity | GO:0002710 |  | 7.343E-05 | 0.03377942 |
|---|
| negative regulation of peptidyl-tyrosine phosphorylation | GO:0050732 |  | 7.343E-05 | 0.02814952 |
|---|
| positive regulation of T cell mediated cytotoxicity | GO:0001916 |  | 7.343E-05 | 0.02412816 |
|---|
| release of sequestered calcium ion into cytosol | GO:0051209 |  | 7.343E-05 | 0.02111214 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 7.343E-05 | 0.01876634 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 8.603E-05 | 0.01978746 |
|---|
| thymic T cell selection | GO:0045061 |  | 1.1E-04 | 0.02299104 |
|---|


