Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 657
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2960 | 7.092e-03 | 3.400e-04 | 3.063e-01 | 7.385e-07 |
|---|
| IPC | 0.4027 | 2.486e-01 | 1.004e-01 | 9.139e-01 | 2.281e-02 |
|---|
| Loi | 0.4392 | 2.870e-04 | 0.000e+00 | 7.476e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6218 | 0.000e+00 | 0.000e+00 | 7.421e-02 | 0.000e+00 |
|---|
| Wang | 0.2394 | 1.520e-02 | 1.016e-01 | 3.131e-01 | 4.839e-04 |
|---|
Expression data for subnetwork 657 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
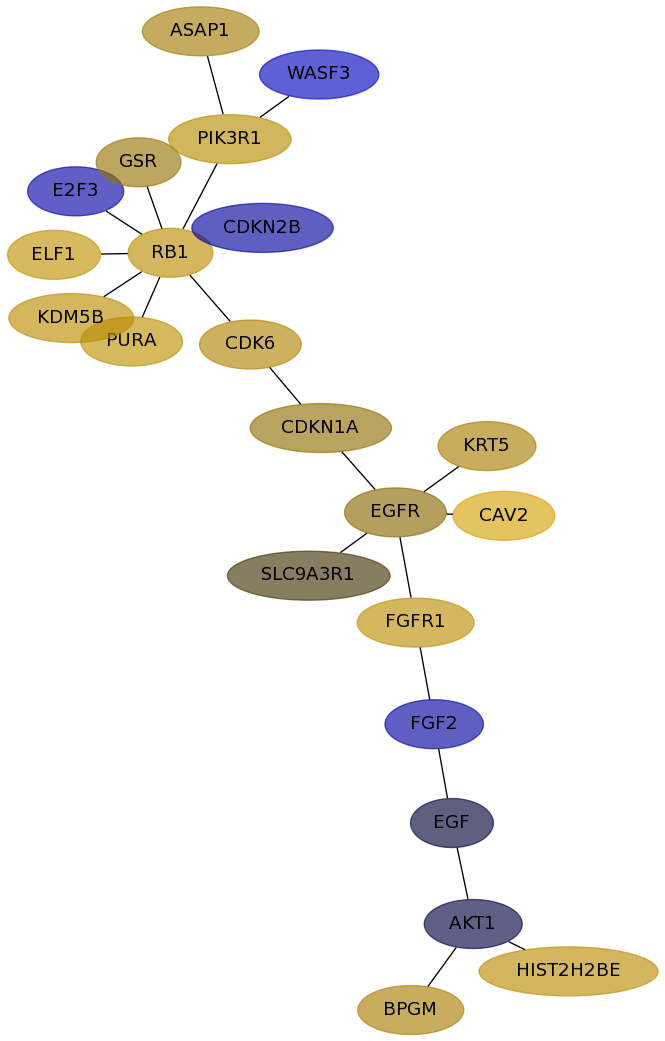
Score for each gene in subnetwork 657 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| CDKN2B |   | 4 | 198 | 937 | 910 | -0.099 | 0.066 | 0.050 | -0.028 | 0.153 |
|---|
| WASF3 |   | 2 | 391 | 1070 | 1062 | -0.213 | 0.038 | -0.040 | 0.036 | 0.054 |
|---|
| KRT5 |   | 5 | 148 | 923 | 898 | 0.129 | -0.125 | 0.046 | 0.064 | -0.190 |
|---|
| KDM5B |   | 3 | 265 | 339 | 348 | 0.199 | -0.011 | 0.279 | -0.047 | 0.161 |
|---|
| AKT1 |   | 13 | 47 | 444 | 430 | -0.012 | 0.045 | 0.126 | -0.099 | -0.168 |
|---|
| FGF2 |   | 1 | 652 | 1070 | 1075 | -0.106 | -0.279 | 0.052 | 0.060 | -0.095 |
|---|
| BPGM |   | 1 | 652 | 1070 | 1075 | 0.127 | 0.243 | -0.056 | -0.170 | 0.186 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| EGF |   | 11 | 59 | 398 | 383 | -0.009 | -0.212 | 0.182 | -0.276 | 0.005 |
|---|
| E2F3 |   | 5 | 148 | 364 | 358 | -0.126 | 0.041 | -0.091 | 0.177 | 0.050 |
|---|
| HIST2H2BE |   | 5 | 148 | 492 | 498 | 0.196 | -0.086 | 0.123 | -0.180 | -0.104 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| GSR |   | 1 | 652 | 1070 | 1075 | 0.087 | -0.018 | -0.011 | -0.194 | -0.126 |
|---|
| SLC9A3R1 |   | 1 | 652 | 1070 | 1075 | 0.011 | 0.004 | 0.017 | 0.017 | -0.110 |
|---|
| ASAP1 |   | 6 | 120 | 129 | 128 | 0.114 | 0.036 | 0.070 | 0.035 | -0.056 |
|---|
| ELF1 |   | 2 | 391 | 1070 | 1062 | 0.229 | -0.076 | 0.083 | -0.101 | -0.077 |
|---|
| PURA |   | 1 | 652 | 1070 | 1075 | 0.220 | 0.049 | 0.177 | -0.061 | 0.090 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
GO Enrichment output for subnetwork 657 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.916E-09 | 6.707E-06 |
|---|
| interphase | GO:0051325 |  | 3.432E-09 | 3.946E-06 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 6.066E-08 | 4.651E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.006E-07 | 5.785E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.152E-07 | 9.901E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.416E-07 | 9.262E-05 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.326E-07 | 2.079E-04 |
|---|
| regulation of granule cell precursor proliferation | GO:0021936 |  | 6.326E-07 | 1.819E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.01E-06 | 2.582E-04 |
|---|
| regulation of nitric-oxide synthase activity | GO:0050999 |  | 2.96E-06 | 6.808E-04 |
|---|
| regulation of monooxygenase activity | GO:0032768 |  | 5.111E-06 | 1.069E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.823E-09 | 4.454E-06 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 4.468E-08 | 5.458E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.004E-08 | 4.075E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.123E-07 | 6.861E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.501E-07 | 7.334E-05 |
|---|
| regulation of granule cell precursor proliferation | GO:0021936 |  | 3.268E-07 | 1.33E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.22E-07 | 1.822E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 7.818E-07 | 2.387E-04 |
|---|
| regulation of nitric-oxide synthase activity | GO:0050999 |  | 1.531E-06 | 4.156E-04 |
|---|
| regulation of monooxygenase activity | GO:0032768 |  | 2.646E-06 | 6.463E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.496E-06 | 7.764E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.093E-09 | 7.442E-06 |
|---|
| interphase | GO:0051325 |  | 3.851E-09 | 4.632E-06 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 6.081E-08 | 4.877E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.611E-08 | 5.18E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.413E-07 | 6.799E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.577E-07 | 1.033E-04 |
|---|
| regulation of granule cell precursor proliferation | GO:0021936 |  | 4.554E-07 | 1.565E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 7.274E-07 | 2.188E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.089E-06 | 2.912E-04 |
|---|
| regulation of nitric-oxide synthase activity | GO:0050999 |  | 2.132E-06 | 5.13E-04 |
|---|
| regulation of monooxygenase activity | GO:0032768 |  | 3.683E-06 | 8.056E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.916E-09 | 6.707E-06 |
|---|
| interphase | GO:0051325 |  | 3.432E-09 | 3.946E-06 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 6.066E-08 | 4.651E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.006E-07 | 5.785E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.152E-07 | 9.901E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.416E-07 | 9.262E-05 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.326E-07 | 2.079E-04 |
|---|
| regulation of granule cell precursor proliferation | GO:0021936 |  | 6.326E-07 | 1.819E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.01E-06 | 2.582E-04 |
|---|
| regulation of nitric-oxide synthase activity | GO:0050999 |  | 2.96E-06 | 6.808E-04 |
|---|
| regulation of monooxygenase activity | GO:0032768 |  | 5.111E-06 | 1.069E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.916E-09 | 6.707E-06 |
|---|
| interphase | GO:0051325 |  | 3.432E-09 | 3.946E-06 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 6.066E-08 | 4.651E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.006E-07 | 5.785E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.152E-07 | 9.901E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.416E-07 | 9.262E-05 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.326E-07 | 2.079E-04 |
|---|
| regulation of granule cell precursor proliferation | GO:0021936 |  | 6.326E-07 | 1.819E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.01E-06 | 2.582E-04 |
|---|
| regulation of nitric-oxide synthase activity | GO:0050999 |  | 2.96E-06 | 6.808E-04 |
|---|
| regulation of monooxygenase activity | GO:0032768 |  | 5.111E-06 | 1.069E-03 |
|---|


