Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 633
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2893 | 9.090e-03 | 5.130e-04 | 3.363e-01 | 1.568e-06 |
|---|
| IPC | 0.4172 | 2.265e-01 | 8.583e-02 | 9.088e-01 | 1.767e-02 |
|---|
| Loi | 0.4376 | 3.070e-04 | 0.000e+00 | 7.582e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6294 | 0.000e+00 | 0.000e+00 | 6.331e-02 | 0.000e+00 |
|---|
| Wang | 0.2397 | 1.501e-02 | 1.008e-01 | 3.113e-01 | 4.710e-04 |
|---|
Expression data for subnetwork 633 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
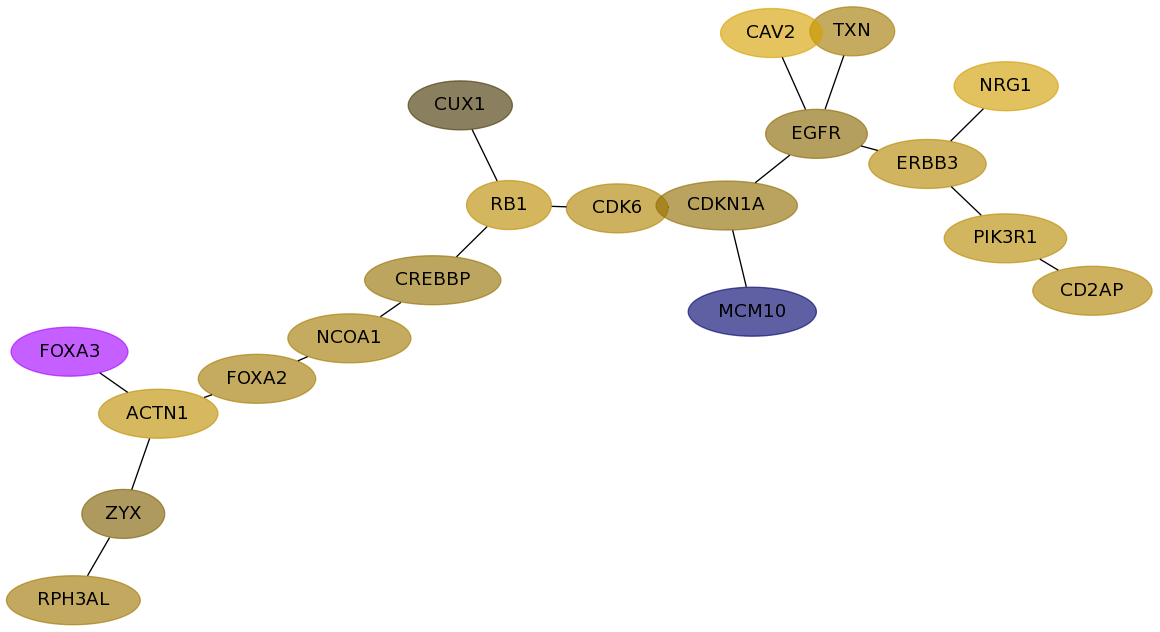
Score for each gene in subnetwork 633 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| MCM10 |   | 3 | 265 | 444 | 447 | -0.033 | 0.178 | -0.245 | 0.015 | -0.063 |
|---|
| TXN |   | 5 | 148 | 657 | 634 | 0.113 | 0.124 | 0.012 | 0.014 | -0.006 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| FOXA3 |   | 6 | 120 | 657 | 631 | undef | 0.192 | 0.289 | undef | undef |
|---|
| CUX1 |   | 8 | 84 | 271 | 266 | 0.012 | -0.042 | 0.256 | -0.121 | 0.040 |
|---|
| ACTN1 |   | 4 | 198 | 657 | 635 | 0.217 | 0.016 | 0.347 | 0.068 | 0.314 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| CREBBP |   | 5 | 148 | 444 | 440 | 0.084 | -0.104 | 0.102 | 0.093 | 0.002 |
|---|
| ZYX |   | 1 | 652 | 657 | 673 | 0.048 | -0.147 | 0.125 | -0.320 | -0.070 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| FOXA2 |   | 1 | 652 | 657 | 673 | 0.117 | 0.000 | -0.024 | -0.078 | 0.153 |
|---|
| CD2AP |   | 1 | 652 | 657 | 673 | 0.154 | 0.156 | 0.104 | -0.086 | 0.117 |
|---|
| RPH3AL |   | 1 | 652 | 657 | 673 | 0.107 | -0.103 | 0.156 | 0.042 | -0.065 |
|---|
| NRG1 |   | 3 | 265 | 262 | 270 | 0.341 | -0.035 | 0.020 | -0.123 | -0.130 |
|---|
| NCOA1 |   | 1 | 652 | 657 | 673 | 0.117 | -0.267 | 0.088 | -0.065 | -0.037 |
|---|
| ERBB3 |   | 5 | 148 | 206 | 202 | 0.175 | -0.171 | -0.007 | -0.141 | -0.098 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
GO Enrichment output for subnetwork 633 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.67E-06 | 3.84E-03 |
|---|
| interphase | GO:0051325 |  | 1.872E-06 | 2.153E-03 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 2.596E-06 | 1.99E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.007E-06 | 1.729E-03 |
|---|
| G1 phase | GO:0051318 |  | 6.564E-06 | 3.02E-03 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 6.564E-06 | 2.516E-03 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 7.958E-06 | 2.615E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 9.399E-06 | 2.702E-03 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 9.534E-06 | 2.436E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.008E-05 | 2.318E-03 |
|---|
| lung development | GO:0030324 |  | 1.079E-05 | 2.257E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 1.673E-10 | 4.088E-07 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 7.066E-10 | 8.631E-07 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 7.066E-10 | 5.754E-07 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 1.137E-09 | 6.944E-07 |
|---|
| lung development | GO:0030324 |  | 5.556E-09 | 2.715E-06 |
|---|
| respiratory tube development | GO:0030323 |  | 7.207E-09 | 2.934E-06 |
|---|
| respiratory system development | GO:0060541 |  | 8.518E-09 | 2.973E-06 |
|---|
| inner ear development | GO:0048839 |  | 7.339E-08 | 2.241E-05 |
|---|
| ear development | GO:0043583 |  | 2.24E-07 | 6.081E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.62E-06 | 3.958E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.238E-06 | 4.971E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 1.716E-10 | 4.129E-07 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 6.051E-10 | 7.28E-07 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 7.929E-10 | 6.359E-07 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 1.024E-09 | 6.162E-07 |
|---|
| lung development | GO:0030324 |  | 7.182E-09 | 3.456E-06 |
|---|
| respiratory tube development | GO:0030323 |  | 9.389E-09 | 3.765E-06 |
|---|
| respiratory system development | GO:0060541 |  | 1.115E-08 | 3.832E-06 |
|---|
| inner ear development | GO:0048839 |  | 7.868E-08 | 2.366E-05 |
|---|
| ear development | GO:0043583 |  | 2.356E-07 | 6.297E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.997E-06 | 4.804E-04 |
|---|
| interphase | GO:0051325 |  | 2.329E-06 | 5.094E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.67E-06 | 3.84E-03 |
|---|
| interphase | GO:0051325 |  | 1.872E-06 | 2.153E-03 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 2.596E-06 | 1.99E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.007E-06 | 1.729E-03 |
|---|
| G1 phase | GO:0051318 |  | 6.564E-06 | 3.02E-03 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 6.564E-06 | 2.516E-03 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 7.958E-06 | 2.615E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 9.399E-06 | 2.702E-03 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 9.534E-06 | 2.436E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.008E-05 | 2.318E-03 |
|---|
| lung development | GO:0030324 |  | 1.079E-05 | 2.257E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.67E-06 | 3.84E-03 |
|---|
| interphase | GO:0051325 |  | 1.872E-06 | 2.153E-03 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 2.596E-06 | 1.99E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.007E-06 | 1.729E-03 |
|---|
| G1 phase | GO:0051318 |  | 6.564E-06 | 3.02E-03 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 6.564E-06 | 2.516E-03 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 7.958E-06 | 2.615E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 9.399E-06 | 2.702E-03 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 9.534E-06 | 2.436E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.008E-05 | 2.318E-03 |
|---|
| lung development | GO:0030324 |  | 1.079E-05 | 2.257E-03 |
|---|


