Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 611
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2834 | 1.124e-02 | 7.310e-04 | 3.636e-01 | 2.988e-06 |
|---|
| IPC | 0.4256 | 2.139e-01 | 7.799e-02 | 9.058e-01 | 1.511e-02 |
|---|
| Loi | 0.4384 | 2.960e-04 | 0.000e+00 | 7.525e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6322 | 0.000e+00 | 0.000e+00 | 5.971e-02 | 0.000e+00 |
|---|
| Wang | 0.2403 | 1.468e-02 | 9.947e-02 | 3.084e-01 | 4.503e-04 |
|---|
Expression data for subnetwork 611 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
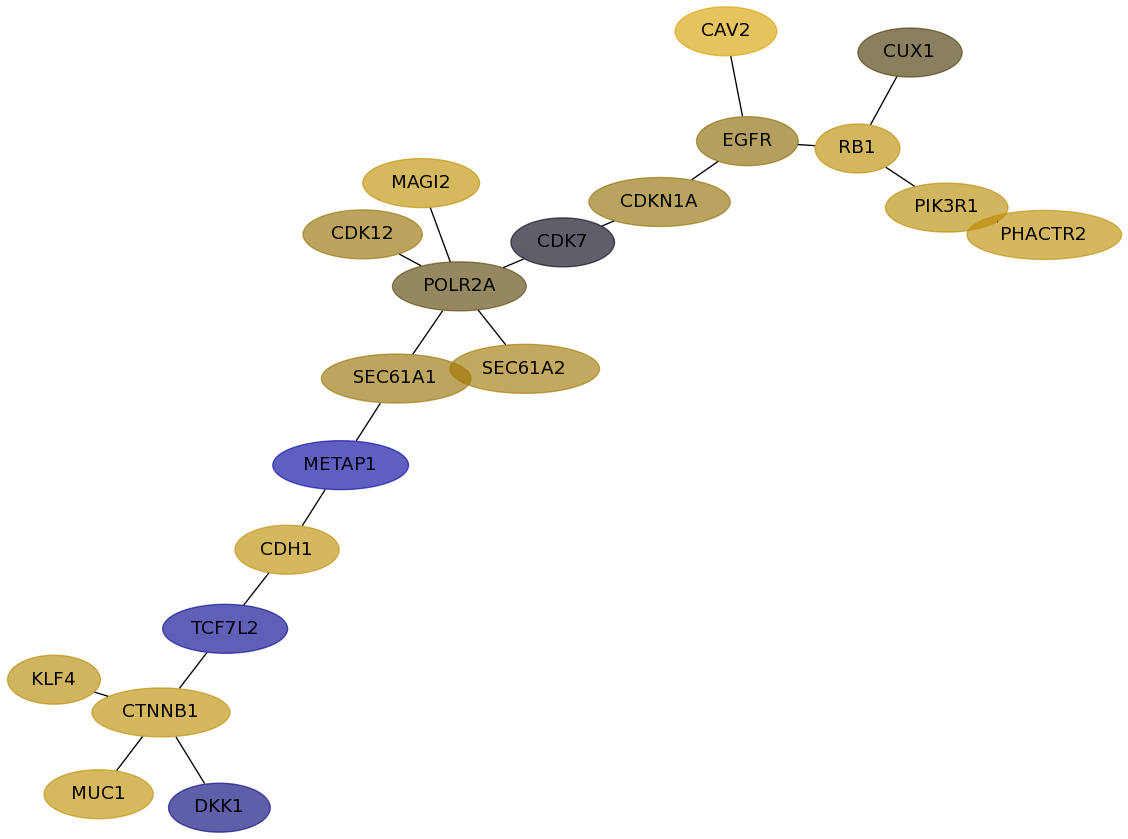
Score for each gene in subnetwork 611 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| CUX1 |   | 8 | 84 | 271 | 266 | 0.012 | -0.042 | 0.256 | -0.121 | 0.040 |
|---|
| PHACTR2 |   | 5 | 148 | 271 | 267 | 0.203 | -0.057 | -0.051 | 0.143 | 0.164 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| CDK7 |   | 4 | 198 | 129 | 130 | -0.002 | 0.105 | 0.184 | 0.017 | -0.097 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| TCF7L2 |   | 17 | 30 | 271 | 264 | -0.071 | -0.021 | 0.143 | 0.280 | 0.101 |
|---|
| POLR2A |   | 5 | 148 | 271 | 267 | 0.019 | -0.107 | 0.247 | 0.091 | 0.141 |
|---|
| DKK1 |   | 4 | 198 | 271 | 273 | -0.041 | 0.134 | 0.091 | -0.124 | 0.129 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| SEC61A1 |   | 5 | 148 | 271 | 267 | 0.085 | -0.161 | 0.128 | -0.096 | 0.070 |
|---|
| METAP1 |   | 1 | 652 | 271 | 311 | -0.102 | 0.017 | -0.015 | 0.080 | 0.005 |
|---|
| KLF4 |   | 3 | 265 | 271 | 280 | 0.174 | 0.035 | 0.060 | -0.026 | -0.025 |
|---|
| MAGI2 |   | 1 | 652 | 271 | 311 | 0.217 | -0.022 | 0.024 | -0.020 | 0.211 |
|---|
| CDK12 |   | 1 | 652 | 271 | 311 | 0.084 | -0.258 | 0.130 | -0.073 | -0.006 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| SEC61A2 |   | 3 | 265 | 271 | 280 | 0.105 | -0.035 | -0.072 | -0.115 | -0.073 |
|---|
GO Enrichment output for subnetwork 611 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 5.704E-07 | 1.312E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 8.258E-07 | 9.497E-04 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 2.227E-06 | 1.707E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.451E-06 | 1.409E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.9E-06 | 2.254E-03 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 5.634E-06 | 2.16E-03 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 6.831E-06 | 2.244E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 7.671E-06 | 2.205E-03 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 8.184E-06 | 2.092E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 8.226E-06 | 1.892E-03 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 8.226E-06 | 1.72E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.454E-11 | 3.553E-08 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 1.929E-10 | 2.357E-07 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 8.145E-10 | 6.633E-07 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 8.145E-10 | 4.975E-07 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 1.31E-09 | 6.403E-07 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.316E-08 | 5.36E-06 |
|---|
| inner ear development | GO:0048839 |  | 8.448E-08 | 2.948E-05 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 1.655E-07 | 5.053E-05 |
|---|
| ear development | GO:0043583 |  | 2.577E-07 | 6.995E-05 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 2.577E-07 | 6.295E-05 |
|---|
| lung development | GO:0030324 |  | 3.225E-07 | 7.163E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.082E-11 | 2.603E-08 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 1.856E-10 | 2.233E-07 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 6.545E-10 | 5.249E-07 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 8.576E-10 | 5.158E-07 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 1.108E-09 | 5.331E-07 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.549E-08 | 6.211E-06 |
|---|
| inner ear development | GO:0048839 |  | 8.502E-08 | 2.922E-05 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 2.001E-07 | 6.018E-05 |
|---|
| ear development | GO:0043583 |  | 2.545E-07 | 6.803E-05 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 2.759E-07 | 6.639E-05 |
|---|
| lung development | GO:0030324 |  | 3.766E-07 | 8.237E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 5.704E-07 | 1.312E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 8.258E-07 | 9.497E-04 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 2.227E-06 | 1.707E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.451E-06 | 1.409E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.9E-06 | 2.254E-03 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 5.634E-06 | 2.16E-03 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 6.831E-06 | 2.244E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 7.671E-06 | 2.205E-03 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 8.184E-06 | 2.092E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 8.226E-06 | 1.892E-03 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 8.226E-06 | 1.72E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 5.704E-07 | 1.312E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 8.258E-07 | 9.497E-04 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 2.227E-06 | 1.707E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.451E-06 | 1.409E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.9E-06 | 2.254E-03 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 5.634E-06 | 2.16E-03 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 6.831E-06 | 2.244E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 7.671E-06 | 2.205E-03 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 8.184E-06 | 2.092E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 8.226E-06 | 1.892E-03 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 8.226E-06 | 1.72E-03 |
|---|


