Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 596
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2785 | 1.338e-02 | 9.760e-04 | 3.868e-01 | 5.050e-06 |
|---|
| IPC | 0.4336 | 2.024e-01 | 7.104e-02 | 9.030e-01 | 1.298e-02 |
|---|
| Loi | 0.4372 | 3.110e-04 | 0.000e+00 | 7.604e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6376 | 0.000e+00 | 0.000e+00 | 5.316e-02 | 0.000e+00 |
|---|
| Wang | 0.2393 | 1.521e-02 | 1.017e-01 | 3.131e-01 | 4.842e-04 |
|---|
Expression data for subnetwork 596 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
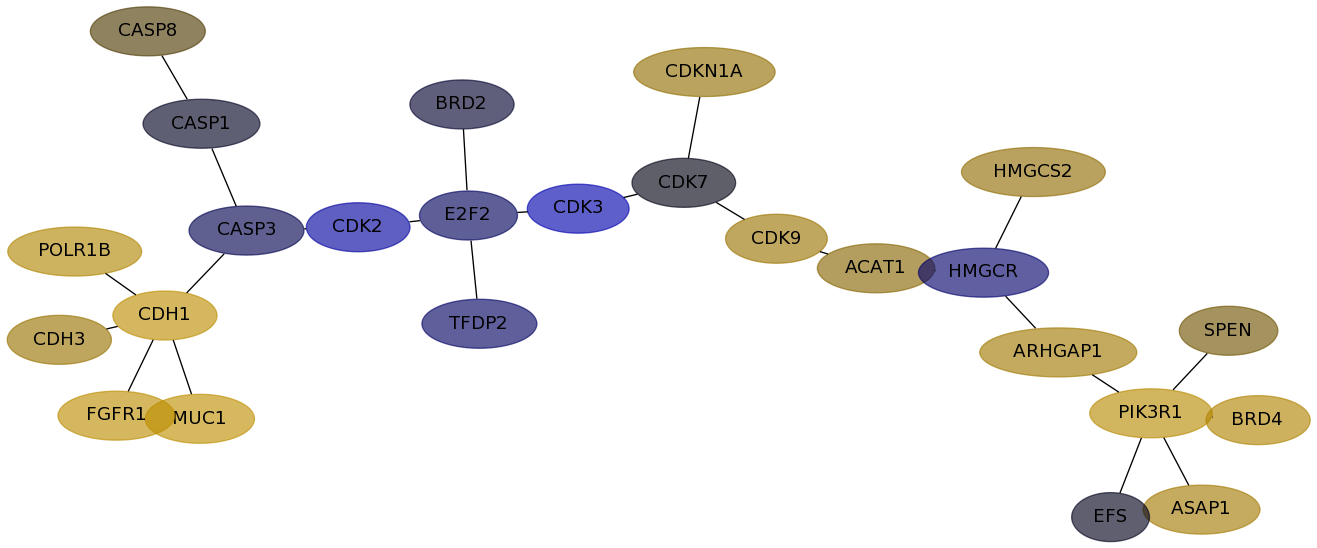
Score for each gene in subnetwork 596 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| CASP3 |   | 4 | 198 | 129 | 130 | -0.015 | 0.234 | -0.032 | -0.142 | -0.084 |
|---|
| BRD4 |   | 18 | 26 | 1 | 12 | 0.156 | -0.119 | 0.095 | -0.075 | -0.044 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| HMGCS2 |   | 3 | 265 | 129 | 139 | 0.073 | -0.277 | 0.226 | -0.194 | -0.085 |
|---|
| EFS |   | 4 | 198 | 129 | 130 | -0.003 | -0.024 | 0.300 | -0.003 | -0.034 |
|---|
| CDK7 |   | 4 | 198 | 129 | 130 | -0.002 | 0.105 | 0.184 | 0.017 | -0.097 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| CDK3 |   | 5 | 148 | 129 | 129 | -0.151 | -0.076 | 0.189 | -0.041 | 0.082 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| BRD2 |   | 3 | 265 | 129 | 139 | -0.006 | 0.016 | 0.348 | 0.105 | 0.093 |
|---|
| ASAP1 |   | 6 | 120 | 129 | 128 | 0.114 | 0.036 | 0.070 | 0.035 | -0.056 |
|---|
| CDK9 |   | 4 | 198 | 129 | 130 | 0.095 | -0.120 | 0.179 | 0.182 | -0.135 |
|---|
| SPEN |   | 3 | 265 | 129 | 139 | 0.034 | -0.147 | 0.207 | -0.086 | 0.238 |
|---|
| ACAT1 |   | 1 | 652 | 129 | 174 | 0.059 | 0.057 | -0.011 | 0.087 | 0.112 |
|---|
| CDH3 |   | 10 | 67 | 1 | 16 | 0.088 | 0.028 | 0.169 | -0.055 | -0.027 |
|---|
| ARHGAP1 |   | 3 | 265 | 129 | 139 | 0.108 | -0.167 | 0.297 | 0.001 | 0.251 |
|---|
| POLR1B |   | 11 | 59 | 129 | 126 | 0.157 | 0.062 | -0.036 | -0.079 | 0.139 |
|---|
| CASP8 |   | 1 | 652 | 129 | 174 | 0.015 | -0.063 | 0.028 | -0.167 | -0.127 |
|---|
| TFDP2 |   | 1 | 652 | 129 | 174 | -0.025 | -0.097 | 0.013 | -0.001 | 0.019 |
|---|
| CDK2 |   | 16 | 34 | 129 | 124 | -0.104 | 0.196 | -0.096 | 0.109 | 0.362 |
|---|
| CASP1 |   | 1 | 652 | 129 | 174 | -0.004 | 0.204 | -0.166 | -0.046 | -0.034 |
|---|
| E2F2 |   | 4 | 198 | 129 | 130 | -0.021 | 0.212 | -0.047 | 0.162 | -0.092 |
|---|
| HMGCR |   | 3 | 265 | 129 | 139 | -0.029 | -0.075 | 0.226 | 0.026 | -0.016 |
|---|
GO Enrichment output for subnetwork 596 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 1.01E-06 | 2.324E-03 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 2.157E-06 | 2.48E-03 |
|---|
| isoprenoid biosynthetic process | GO:0008299 |  | 8.101E-06 | 6.211E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.044E-05 | 6.006E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.632E-05 | 7.505E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.749E-05 | 6.704E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.139E-05 | 7.027E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.341E-05 | 6.731E-03 |
|---|
| cholesterol biosynthetic process | GO:0006695 |  | 2.341E-05 | 5.983E-03 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 3.692E-05 | 8.491E-03 |
|---|
| sterol biosynthetic process | GO:0016126 |  | 5.691E-05 | 0.0119001 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 9.871E-07 | 2.411E-03 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 1.804E-06 | 2.204E-03 |
|---|
| isoprenoid biosynthetic process | GO:0008299 |  | 4.566E-06 | 3.718E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.955E-06 | 3.637E-03 |
|---|
| cholesterol biosynthetic process | GO:0006695 |  | 1.244E-05 | 6.08E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.279E-05 | 5.209E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.355E-05 | 4.728E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 1.69E-05 | 5.159E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.85E-05 | 5.023E-03 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 2.417E-05 | 5.904E-03 |
|---|
| sterol biosynthetic process | GO:0016126 |  | 3.242E-05 | 7.201E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 1.249E-06 | 3.004E-03 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 2.282E-06 | 2.745E-03 |
|---|
| isoprenoid biosynthetic process | GO:0008299 |  | 4.697E-06 | 3.767E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.084E-06 | 4.863E-03 |
|---|
| cholesterol biosynthetic process | GO:0006695 |  | 1.36E-05 | 6.544E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.543E-05 | 6.186E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.636E-05 | 5.624E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.053E-05 | 6.176E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.337E-05 | 6.247E-03 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 2.819E-05 | 6.782E-03 |
|---|
| sterol biosynthetic process | GO:0016126 |  | 3.689E-05 | 8.069E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 1.01E-06 | 2.324E-03 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 2.157E-06 | 2.48E-03 |
|---|
| isoprenoid biosynthetic process | GO:0008299 |  | 8.101E-06 | 6.211E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.044E-05 | 6.006E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.632E-05 | 7.505E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.749E-05 | 6.704E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.139E-05 | 7.027E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.341E-05 | 6.731E-03 |
|---|
| cholesterol biosynthetic process | GO:0006695 |  | 2.341E-05 | 5.983E-03 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 3.692E-05 | 8.491E-03 |
|---|
| sterol biosynthetic process | GO:0016126 |  | 5.691E-05 | 0.0119001 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 1.01E-06 | 2.324E-03 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 2.157E-06 | 2.48E-03 |
|---|
| isoprenoid biosynthetic process | GO:0008299 |  | 8.101E-06 | 6.211E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.044E-05 | 6.006E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.632E-05 | 7.505E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.749E-05 | 6.704E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.139E-05 | 7.027E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.341E-05 | 6.731E-03 |
|---|
| cholesterol biosynthetic process | GO:0006695 |  | 2.341E-05 | 5.983E-03 |
|---|
| transcription initiation from RNA polymerase II promoter | GO:0006367 |  | 3.692E-05 | 8.491E-03 |
|---|
| sterol biosynthetic process | GO:0016126 |  | 5.691E-05 | 0.0119001 |
|---|


