Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 578
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2747 | 1.523e-02 | 1.210e-03 | 4.047e-01 | 7.456e-06 |
|---|
| IPC | 0.4423 | 1.900e-01 | 6.394e-02 | 9.000e-01 | 1.093e-02 |
|---|
| Loi | 0.4335 | 3.630e-04 | 0.000e+00 | 7.852e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6380 | 0.000e+00 | 0.000e+00 | 5.262e-02 | 0.000e+00 |
|---|
| Wang | 0.2389 | 1.547e-02 | 1.027e-01 | 3.155e-01 | 5.015e-04 |
|---|
Expression data for subnetwork 578 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
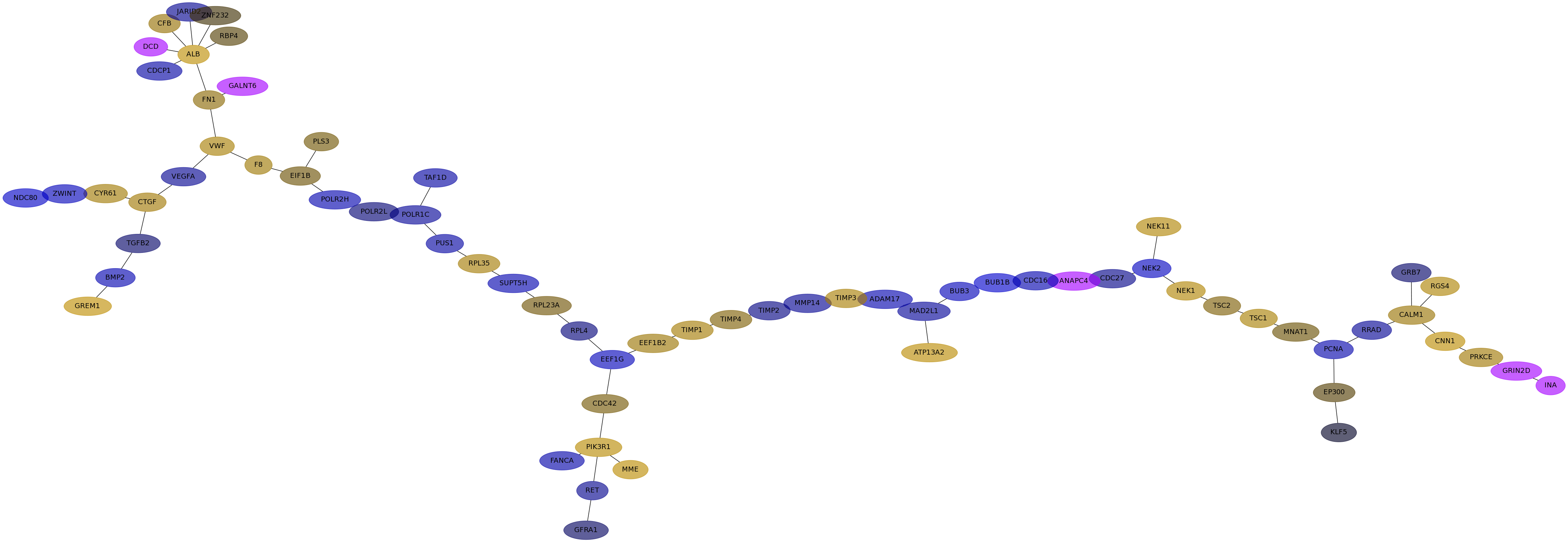
Score for each gene in subnetwork 578 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| RGS4 |   | 3 | 265 | 57 | 71 | 0.144 | 0.022 | 0.160 | -0.006 | 0.371 |
|---|
| CDCP1 |   | 2 | 391 | 57 | 80 | -0.117 | 0.068 | 0.045 | -0.058 | 0.010 |
|---|
| TSC2 |   | 3 | 265 | 57 | 71 | 0.046 | 0.236 | 0.044 | 0.114 | -0.102 |
|---|
| GALNT6 |   | 1 | 652 | 57 | 94 | undef | 0.009 | 0.114 | undef | undef |
|---|
| RET |   | 4 | 198 | 57 | 68 | -0.069 | 0.098 | 0.063 | -0.179 | -0.159 |
|---|
| NDC80 |   | 2 | 391 | 57 | 80 | -0.266 | 0.246 | -0.229 | 0.060 | 0.044 |
|---|
| RRAD |   | 1 | 652 | 57 | 94 | -0.079 | -0.081 | 0.171 | -0.082 | 0.117 |
|---|
| EEF1G |   | 1 | 652 | 57 | 94 | -0.197 | 0.116 | -0.224 | 0.131 | 0.082 |
|---|
| EIF1B |   | 6 | 120 | 57 | 62 | 0.029 | 0.149 | 0.075 | 0.332 | -0.070 |
|---|
| PLS3 |   | 2 | 391 | 57 | 80 | 0.034 | 0.144 | 0.175 | 0.247 | 0.256 |
|---|
| RPL23A |   | 10 | 67 | 57 | 52 | 0.031 | 0.208 | -0.007 | 0.024 | 0.055 |
|---|
| EEF1B2 |   | 1 | 652 | 57 | 94 | 0.092 | 0.143 | -0.071 | 0.141 | -0.026 |
|---|
| PCNA |   | 3 | 265 | 57 | 71 | -0.138 | 0.243 | -0.158 | 0.027 | 0.003 |
|---|
| CTGF |   | 10 | 67 | 57 | 52 | 0.106 | -0.169 | 0.181 | 0.126 | 0.209 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| DCD |   | 2 | 391 | 57 | 80 | undef | 0.135 | undef | undef | undef |
|---|
| MMP14 |   | 2 | 391 | 57 | 80 | -0.077 | -0.114 | 0.056 | 0.011 | 0.148 |
|---|
| RPL35 |   | 5 | 148 | 57 | 63 | 0.114 | 0.108 | -0.012 | 0.370 | -0.220 |
|---|
| TIMP2 |   | 3 | 265 | 1 | 24 | -0.053 | -0.126 | 0.212 | 0.021 | 0.137 |
|---|
| POLR2L |   | 3 | 265 | 57 | 71 | -0.038 | 0.037 | -0.012 | 0.224 | 0.015 |
|---|
| BUB1B |   | 2 | 391 | 57 | 80 | -0.289 | 0.050 | -0.164 | 0.037 | 0.148 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| INA |   | 2 | 391 | 57 | 80 | undef | 0.144 | 0.025 | undef | undef |
|---|
| ATP13A2 |   | 1 | 652 | 57 | 94 | 0.193 | -0.111 | 0.043 | -0.240 | -0.042 |
|---|
| RBP4 |   | 1 | 652 | 57 | 94 | 0.017 | -0.059 | -0.011 | 0.206 | -0.045 |
|---|
| ZNF232 |   | 5 | 148 | 57 | 63 | 0.010 | 0.048 | -0.040 | 0.189 | 0.159 |
|---|
| CDC16 |   | 1 | 652 | 57 | 94 | -0.152 | 0.086 | -0.029 | 0.095 | 0.022 |
|---|
| MAD2L1 |   | 2 | 391 | 57 | 80 | -0.089 | 0.208 | -0.130 | 0.116 | 0.063 |
|---|
| POLR2H |   | 2 | 391 | 57 | 80 | -0.150 | 0.255 | -0.016 | 0.147 | 0.108 |
|---|
| KLF5 |   | 25 | 19 | 57 | 50 | -0.005 | 0.077 | 0.117 | -0.007 | -0.006 |
|---|
| PUS1 |   | 1 | 652 | 57 | 94 | -0.112 | -0.026 | 0.045 | 0.044 | 0.065 |
|---|
| FN1 |   | 5 | 148 | 57 | 63 | 0.064 | 0.190 | 0.133 | 0.091 | 0.083 |
|---|
| F8 |   | 2 | 391 | 57 | 80 | 0.102 | 0.059 | -0.037 | 0.088 | 0.039 |
|---|
| TIMP4 |   | 1 | 652 | 57 | 94 | 0.047 | 0.050 | 0.044 | 0.193 | -0.153 |
|---|
| TGFB2 |   | 7 | 100 | 57 | 60 | -0.028 | -0.063 | 0.244 | 0.226 | 0.015 |
|---|
| PRKCE |   | 1 | 652 | 57 | 94 | 0.107 | -0.166 | 0.241 | -0.088 | -0.101 |
|---|
| BMP2 |   | 7 | 100 | 33 | 37 | -0.155 | -0.011 | 0.097 | 0.148 | -0.069 |
|---|
| TIMP1 |   | 1 | 652 | 57 | 94 | 0.116 | 0.010 | 0.010 | 0.143 | 0.059 |
|---|
| CYR61 |   | 2 | 391 | 57 | 80 | 0.109 | 0.018 | 0.206 | 0.203 | 0.234 |
|---|
| NEK2 |   | 1 | 652 | 57 | 94 | -0.222 | 0.292 | -0.106 | 0.020 | 0.165 |
|---|
| GFRA1 |   | 2 | 391 | 57 | 80 | -0.023 | 0.011 | -0.067 | 0.292 | 0.029 |
|---|
| GREM1 |   | 5 | 148 | 57 | 63 | 0.213 | -0.073 | 0.200 | 0.028 | 0.261 |
|---|
| ZWINT |   | 1 | 652 | 57 | 94 | -0.196 | 0.188 | -0.115 | 0.037 | -0.004 |
|---|
| ADAM17 |   | 1 | 652 | 57 | 94 | -0.151 | -0.113 | 0.058 | -0.073 | -0.106 |
|---|
| BUB3 |   | 1 | 652 | 57 | 94 | -0.187 | 0.133 | -0.053 | 0.165 | 0.181 |
|---|
| TAF1D |   | 5 | 148 | 57 | 63 | -0.110 | 0.141 | 0.113 | 0.041 | 0.059 |
|---|
| CALM1 |   | 3 | 265 | 57 | 71 | 0.091 | 0.014 | 0.021 | 0.192 | 0.247 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| SUPT5H |   | 9 | 77 | 1 | 18 | -0.150 | -0.085 | 0.007 | -0.298 | -0.068 |
|---|
| FANCA |   | 3 | 265 | 57 | 71 | -0.131 | 0.120 | -0.188 | 0.106 | -0.053 |
|---|
| CNN1 |   | 1 | 652 | 57 | 94 | 0.201 | -0.190 | 0.240 | 0.209 | 0.173 |
|---|
| MNAT1 |   | 2 | 391 | 57 | 80 | 0.029 | 0.194 | 0.055 | 0.224 | 0.236 |
|---|
| TSC1 |   | 1 | 652 | 57 | 94 | 0.132 | -0.094 | 0.257 | 0.103 | 0.077 |
|---|
| TIMP3 |   | 1 | 652 | 57 | 94 | 0.127 | 0.007 | 0.205 | 0.080 | 0.157 |
|---|
| NEK1 |   | 4 | 198 | 57 | 68 | 0.155 | 0.032 | 0.184 | 0.049 | 0.135 |
|---|
| CDC27 |   | 4 | 198 | 1 | 23 | -0.062 | 0.156 | -0.086 | -0.029 | 0.348 |
|---|
| ALB |   | 7 | 100 | 57 | 60 | 0.207 | -0.122 | 0.218 | 0.080 | 0.038 |
|---|
| GRB7 |   | 13 | 47 | 57 | 51 | -0.028 | 0.021 | 0.115 | -0.072 | 0.129 |
|---|
| NEK11 |   | 1 | 652 | 57 | 94 | 0.153 | 0.081 | 0.152 | -0.197 | 0.040 |
|---|
| VWF |   | 4 | 198 | 57 | 68 | 0.126 | -0.091 | 0.072 | 0.152 | 0.100 |
|---|
| GRIN2D |   | 18 | 26 | 33 | 32 | undef | 0.058 | 0.155 | 0.000 | undef |
|---|
| CFB |   | 1 | 652 | 57 | 94 | 0.081 | 0.138 | 0.046 | 0.109 | -0.149 |
|---|
| RPL4 |   | 1 | 652 | 57 | 94 | -0.042 | 0.057 | 0.095 | 0.248 | -0.014 |
|---|
| POLR1C |   | 2 | 391 | 57 | 80 | -0.086 | 0.176 | -0.210 | 0.181 | 0.038 |
|---|
| MME |   | 8 | 84 | 57 | 54 | 0.200 | -0.121 | 0.020 | 0.088 | -0.130 |
|---|
| ANAPC4 |   | 1 | 652 | 57 | 94 | undef | 0.071 | 0.091 | undef | undef |
|---|
| JARID2 |   | 1 | 652 | 57 | 94 | -0.073 | -0.000 | 0.044 | 0.136 | -0.099 |
|---|
GO Enrichment output for subnetwork 578 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| tube morphogenesis | GO:0035239 |  | 8.89E-07 | 2.045E-03 |
|---|
| nuclear migration | GO:0007097 |  | 3.532E-06 | 4.061E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.532E-06 | 2.708E-03 |
|---|
| glial cell migration | GO:0008347 |  | 3.532E-06 | 2.031E-03 |
|---|
| mitotic cell cycle checkpoint | GO:0007093 |  | 4.064E-06 | 1.87E-03 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 4.324E-06 | 1.658E-03 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 4.768E-06 | 1.567E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 4.768E-06 | 1.371E-03 |
|---|
| angiogenesis | GO:0001525 |  | 5.295E-06 | 1.353E-03 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 6.327E-06 | 1.455E-03 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 6.929E-06 | 1.449E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 2.586E-08 | 6.318E-05 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 2.872E-08 | 3.508E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.872E-08 | 2.339E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 4.298E-08 | 2.625E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 8.232E-08 | 4.022E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.262E-07 | 5.138E-05 |
|---|
| tube morphogenesis | GO:0035239 |  | 1.883E-07 | 6.572E-05 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 2.55E-07 | 7.786E-05 |
|---|
| glial cell migration | GO:0008347 |  | 9.404E-07 | 2.553E-04 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 9.493E-07 | 2.319E-04 |
|---|
| mitotic cell cycle checkpoint | GO:0007093 |  | 9.679E-07 | 2.15E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 5.125E-08 | 1.233E-04 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 5.689E-08 | 6.843E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 5.689E-08 | 4.562E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 8.499E-08 | 5.112E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.623E-07 | 7.812E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 2.483E-07 | 9.959E-05 |
|---|
| tube morphogenesis | GO:0035239 |  | 2.921E-07 | 1.004E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 5E-07 | 1.504E-04 |
|---|
| glial cell migration | GO:0008347 |  | 1.276E-06 | 3.411E-04 |
|---|
| mitotic cell cycle checkpoint | GO:0007093 |  | 1.389E-06 | 3.343E-04 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 1.434E-06 | 3.137E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| tube morphogenesis | GO:0035239 |  | 8.89E-07 | 2.045E-03 |
|---|
| nuclear migration | GO:0007097 |  | 3.532E-06 | 4.061E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.532E-06 | 2.708E-03 |
|---|
| glial cell migration | GO:0008347 |  | 3.532E-06 | 2.031E-03 |
|---|
| mitotic cell cycle checkpoint | GO:0007093 |  | 4.064E-06 | 1.87E-03 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 4.324E-06 | 1.658E-03 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 4.768E-06 | 1.567E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 4.768E-06 | 1.371E-03 |
|---|
| angiogenesis | GO:0001525 |  | 5.295E-06 | 1.353E-03 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 6.327E-06 | 1.455E-03 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 6.929E-06 | 1.449E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| tube morphogenesis | GO:0035239 |  | 8.89E-07 | 2.045E-03 |
|---|
| nuclear migration | GO:0007097 |  | 3.532E-06 | 4.061E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.532E-06 | 2.708E-03 |
|---|
| glial cell migration | GO:0008347 |  | 3.532E-06 | 2.031E-03 |
|---|
| mitotic cell cycle checkpoint | GO:0007093 |  | 4.064E-06 | 1.87E-03 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 4.324E-06 | 1.658E-03 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 4.768E-06 | 1.567E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 4.768E-06 | 1.371E-03 |
|---|
| angiogenesis | GO:0001525 |  | 5.295E-06 | 1.353E-03 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 6.327E-06 | 1.455E-03 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 6.929E-06 | 1.449E-03 |
|---|


