Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 574
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2759 | 1.461e-02 | 1.130e-03 | 3.990e-01 | 6.588e-06 |
|---|
| IPC | 0.4415 | 1.912e-01 | 6.462e-02 | 9.003e-01 | 1.112e-02 |
|---|
| Loi | 0.4338 | 3.590e-04 | 0.000e+00 | 7.834e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6368 | 0.000e+00 | 0.000e+00 | 5.404e-02 | 0.000e+00 |
|---|
| Wang | 0.2370 | 1.658e-02 | 1.072e-01 | 3.251e-01 | 5.777e-04 |
|---|
Expression data for subnetwork 574 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
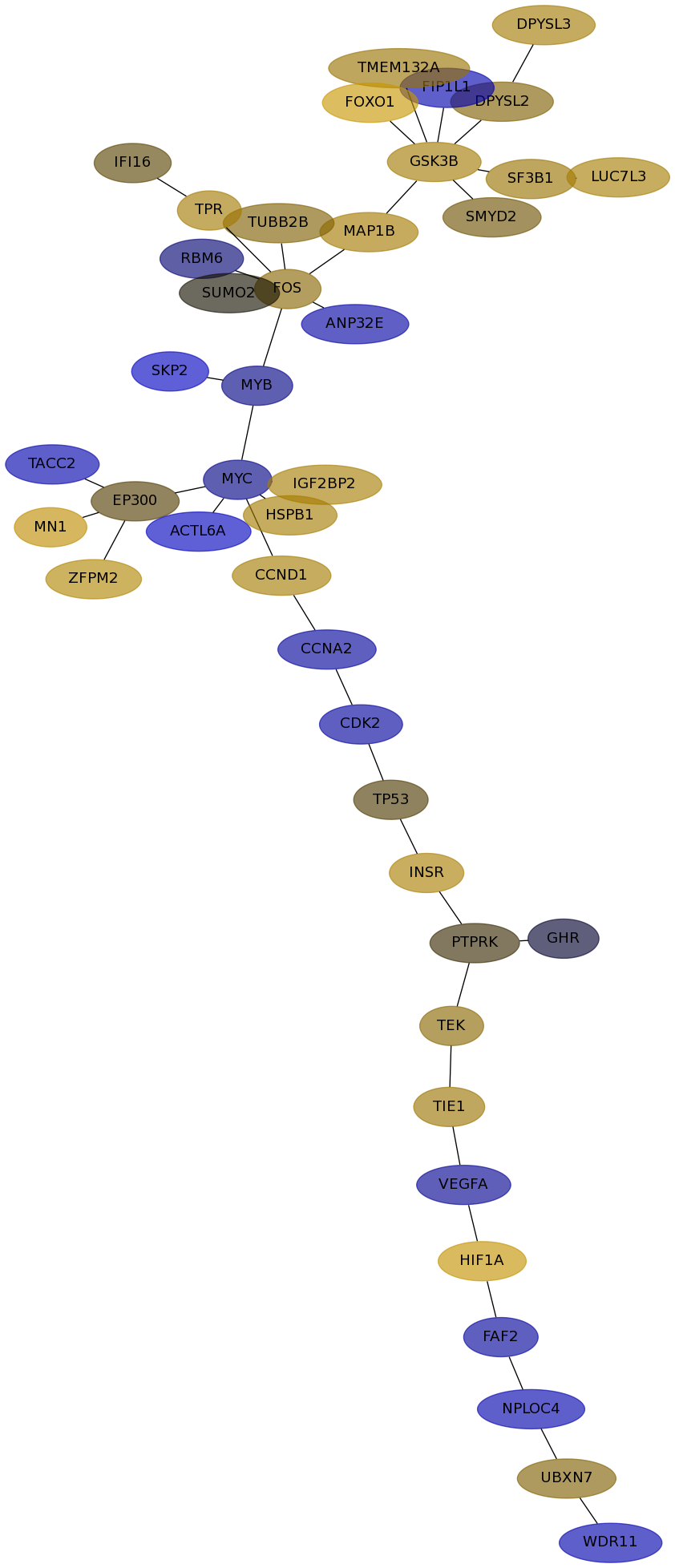
Score for each gene in subnetwork 574 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| TEK |   | 7 | 100 | 148 | 155 | 0.063 | -0.149 | -0.049 | 0.174 | 0.113 |
|---|
| FAF2 |   | 2 | 391 | 148 | 171 | -0.093 | 0.027 | 0.212 | -0.070 | 0.195 |
|---|
| FOXO1 |   | 4 | 198 | 148 | 162 | 0.278 | 0.072 | 0.094 | -0.021 | 0.221 |
|---|
| SF3B1 |   | 15 | 40 | 148 | 144 | 0.091 | -0.154 | 0.126 | -0.073 | -0.204 |
|---|
| SUMO2 |   | 12 | 55 | 148 | 150 | 0.003 | 0.117 | 0.062 | 0.294 | -0.096 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| IGF2BP2 |   | 3 | 265 | 148 | 166 | 0.124 | 0.210 | -0.002 | 0.098 | -0.122 |
|---|
| TMEM132A |   | 7 | 100 | 33 | 37 | 0.088 | 0.019 | 0.127 | -0.141 | -0.092 |
|---|
| NPLOC4 |   | 2 | 391 | 148 | 171 | -0.146 | -0.237 | -0.168 | 0.106 | 0.019 |
|---|
| UBXN7 |   | 6 | 120 | 148 | 156 | 0.049 | -0.055 | 0.153 | 0.224 | 0.175 |
|---|
| CCND1 |   | 12 | 55 | 148 | 150 | 0.122 | -0.258 | 0.074 | 0.117 | 0.003 |
|---|
| CCNA2 |   | 15 | 40 | 148 | 144 | -0.097 | 0.176 | -0.053 | 0.188 | 0.246 |
|---|
| TACC2 |   | 9 | 77 | 148 | 154 | -0.151 | -0.073 | 0.122 | 0.181 | 0.180 |
|---|
| MYC |   | 24 | 21 | 148 | 137 | -0.056 | -0.101 | -0.169 | 0.188 | -0.134 |
|---|
| DPYSL2 |   | 3 | 265 | 148 | 166 | 0.050 | -0.095 | -0.037 | 0.296 | 0.104 |
|---|
| GHR |   | 14 | 42 | 148 | 146 | -0.006 | -0.112 | 0.174 | 0.090 | 0.201 |
|---|
| RBM6 |   | 10 | 67 | 148 | 152 | -0.036 | -0.082 | 0.265 | -0.004 | 0.111 |
|---|
| MYB |   | 3 | 265 | 148 | 166 | -0.054 | -0.028 | 0.075 | 0.002 | -0.012 |
|---|
| PTPRK |   | 6 | 120 | 148 | 156 | 0.008 | 0.156 | 0.141 | -0.094 | -0.024 |
|---|
| MN1 |   | 6 | 120 | 148 | 156 | 0.213 | -0.166 | -0.047 | 0.150 | 0.011 |
|---|
| SKP2 |   | 14 | 42 | 148 | 146 | -0.228 | 0.092 | -0.043 | 0.237 | -0.009 |
|---|
| ZFPM2 |   | 4 | 198 | 148 | 162 | 0.160 | 0.001 | 0.092 | 0.113 | 0.379 |
|---|
| SMYD2 |   | 4 | 198 | 148 | 162 | 0.032 | 0.260 | 0.068 | 0.179 | 0.191 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| FOS |   | 20 | 24 | 148 | 138 | 0.060 | -0.137 | 0.129 | 0.185 | 0.234 |
|---|
| MAP1B |   | 13 | 47 | 148 | 148 | 0.118 | -0.123 | 0.263 | -0.107 | 0.100 |
|---|
| DPYSL3 |   | 2 | 391 | 148 | 171 | 0.113 | -0.065 | 0.133 | 0.021 | 0.261 |
|---|
| TPR |   | 10 | 67 | 148 | 152 | 0.118 | -0.130 | 0.120 | -0.036 | 0.156 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| WDR11 |   | 1 | 652 | 148 | 185 | -0.145 | -0.032 | -0.033 | 0.186 | 0.162 |
|---|
| HIF1A |   | 21 | 23 | 33 | 31 | 0.246 | -0.098 | 0.224 | -0.118 | 0.356 |
|---|
| LUC7L3 |   | 13 | 47 | 148 | 148 | 0.123 | 0.043 | 0.237 | 0.175 | 0.074 |
|---|
| TUBB2B |   | 5 | 148 | 148 | 161 | 0.048 | -0.036 | 0.304 | 0.045 | 0.139 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| TIE1 |   | 6 | 120 | 148 | 156 | 0.094 | -0.108 | 0.005 | 0.141 | 0.252 |
|---|
| ANP32E |   | 6 | 120 | 148 | 156 | -0.114 | 0.166 | -0.139 | 0.191 | 0.118 |
|---|
| FIP1L1 |   | 1 | 652 | 148 | 185 | -0.146 | 0.173 | -0.002 | 0.204 | -0.116 |
|---|
| CDK2 |   | 16 | 34 | 129 | 124 | -0.104 | 0.196 | -0.096 | 0.109 | 0.362 |
|---|
| HSPB1 |   | 4 | 198 | 148 | 162 | 0.121 | 0.053 | 0.169 | 0.017 | 0.161 |
|---|
| ACTL6A |   | 1 | 652 | 148 | 185 | -0.214 | 0.095 | -0.069 | 0.193 | 0.040 |
|---|
| IFI16 |   | 1 | 652 | 148 | 185 | 0.021 | 0.134 | -0.026 | 0.110 | -0.051 |
|---|
GO Enrichment output for subnetwork 574 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 7.538E-07 | 1.734E-03 |
|---|
| ER overload response | GO:0006983 |  | 1.503E-06 | 1.728E-03 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 1.503E-06 | 1.152E-03 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.701E-06 | 9.782E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.769E-06 | 8.138E-04 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.882E-06 | 7.216E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 2.557E-06 | 8.4E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 2.601E-06 | 7.477E-04 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 2.601E-06 | 6.646E-04 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 2.886E-06 | 6.637E-04 |
|---|
| peptidyl-tyrosine modification | GO:0018212 |  | 3.691E-06 | 7.718E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 2.494E-07 | 6.093E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 4.571E-07 | 5.584E-04 |
|---|
| regulation of DNA replication | GO:0006275 |  | 6.924E-07 | 5.639E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 8.267E-07 | 5.049E-04 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 1.022E-06 | 4.995E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.022E-06 | 4.162E-04 |
|---|
| ER overload response | GO:0006983 |  | 1.388E-06 | 4.843E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 1.388E-06 | 4.238E-04 |
|---|
| peptidyl-tyrosine modification | GO:0018212 |  | 1.394E-06 | 3.783E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.466E-06 | 3.582E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.613E-06 | 3.583E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 3.325E-07 | 8E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 7.912E-07 | 9.518E-04 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.102E-06 | 8.839E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.205E-06 | 7.248E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.265E-06 | 6.088E-04 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 1.491E-06 | 5.981E-04 |
|---|
| regulation of homeostatic process | GO:0032844 |  | 1.839E-06 | 6.319E-04 |
|---|
| ER overload response | GO:0006983 |  | 1.849E-06 | 5.561E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 1.849E-06 | 4.943E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 2.032E-06 | 4.89E-04 |
|---|
| peptidyl-tyrosine modification | GO:0018212 |  | 2.032E-06 | 4.446E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 7.538E-07 | 1.734E-03 |
|---|
| ER overload response | GO:0006983 |  | 1.503E-06 | 1.728E-03 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 1.503E-06 | 1.152E-03 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.701E-06 | 9.782E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.769E-06 | 8.138E-04 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.882E-06 | 7.216E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 2.557E-06 | 8.4E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 2.601E-06 | 7.477E-04 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 2.601E-06 | 6.646E-04 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 2.886E-06 | 6.637E-04 |
|---|
| peptidyl-tyrosine modification | GO:0018212 |  | 3.691E-06 | 7.718E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 7.538E-07 | 1.734E-03 |
|---|
| ER overload response | GO:0006983 |  | 1.503E-06 | 1.728E-03 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 1.503E-06 | 1.152E-03 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.701E-06 | 9.782E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 1.769E-06 | 8.138E-04 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.882E-06 | 7.216E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 2.557E-06 | 8.4E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 2.601E-06 | 7.477E-04 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 2.601E-06 | 6.646E-04 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 2.886E-06 | 6.637E-04 |
|---|
| peptidyl-tyrosine modification | GO:0018212 |  | 3.691E-06 | 7.718E-04 |
|---|


