Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 562
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2789 | 1.319e-02 | 9.530e-04 | 3.849e-01 | 4.838e-06 |
|---|
| IPC | 0.4420 | 1.905e-01 | 6.422e-02 | 9.001e-01 | 1.101e-02 |
|---|
| Loi | 0.4323 | 3.810e-04 | 0.000e+00 | 7.933e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6310 | 0.000e+00 | 0.000e+00 | 6.126e-02 | 0.000e+00 |
|---|
| Wang | 0.2346 | 1.801e-02 | 1.128e-01 | 3.369e-01 | 6.847e-04 |
|---|
Expression data for subnetwork 562 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
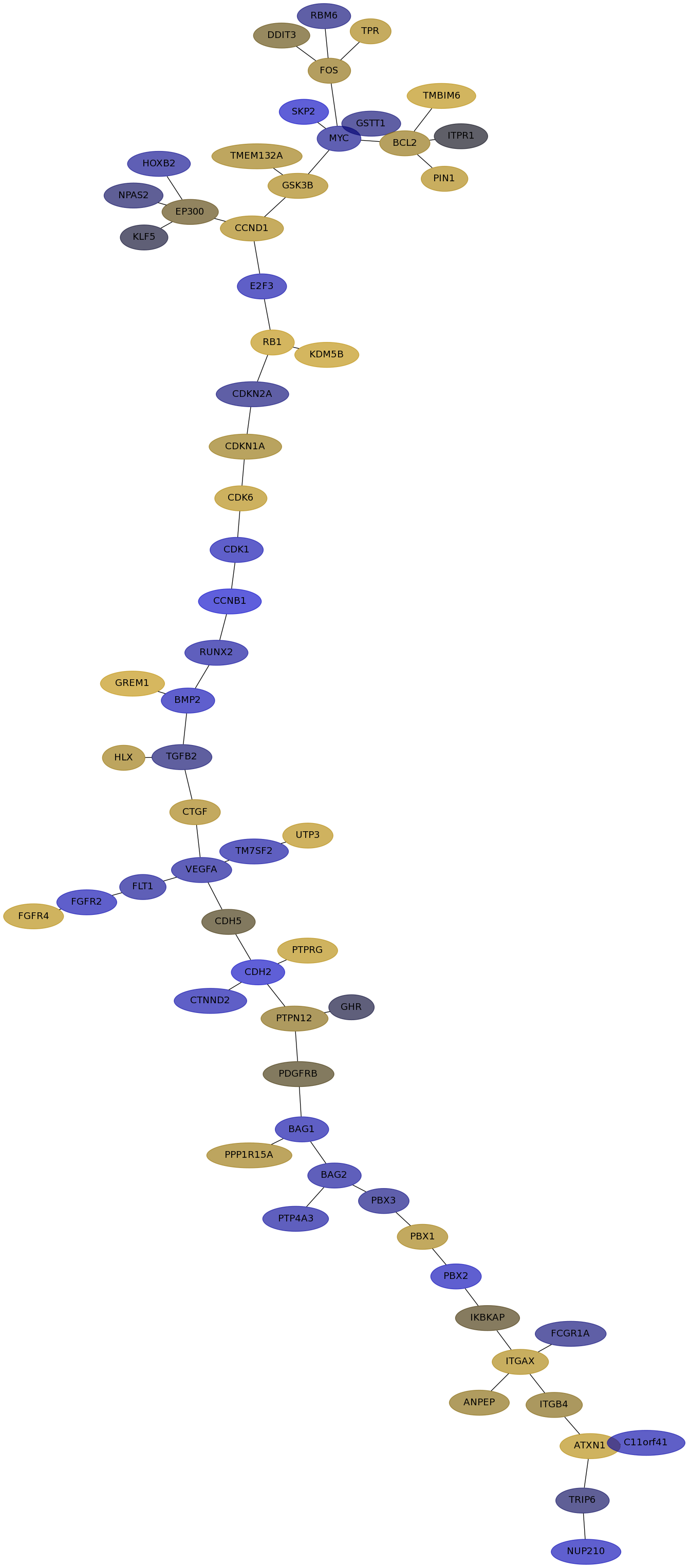
Score for each gene in subnetwork 562 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| RUNX2 |   | 2 | 391 | 364 | 375 | -0.087 | 0.184 | -0.104 | -0.127 | undef |
|---|
| TRIP6 |   | 3 | 265 | 364 | 362 | -0.020 | 0.170 | 0.145 | 0.034 | -0.213 |
|---|
| NPAS2 |   | 3 | 265 | 364 | 362 | -0.019 | 0.116 | 0.253 | -0.157 | -0.093 |
|---|
| C11orf41 |   | 2 | 391 | 364 | 375 | -0.126 | 0.062 | 0.058 | 0.045 | -0.157 |
|---|
| PBX3 |   | 1 | 652 | 364 | 388 | -0.045 | 0.065 | 0.008 | 0.034 | 0.021 |
|---|
| GSTT1 |   | 6 | 120 | 364 | 355 | -0.034 | 0.148 | 0.107 | -0.190 | -0.064 |
|---|
| PTP4A3 |   | 3 | 265 | 364 | 362 | -0.090 | 0.137 | -0.196 | 0.085 | -0.004 |
|---|
| PBX1 |   | 1 | 652 | 364 | 388 | 0.103 | -0.010 | 0.145 | -0.013 | -0.053 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| TGFB2 |   | 7 | 100 | 57 | 60 | -0.028 | -0.063 | 0.244 | 0.226 | 0.015 |
|---|
| IKBKAP |   | 5 | 148 | 364 | 358 | 0.010 | 0.089 | 0.184 | 0.142 | -0.017 |
|---|
| BAG2 |   | 2 | 391 | 364 | 375 | -0.081 | 0.016 | 0.178 | 0.152 | 0.083 |
|---|
| PTPRG |   | 1 | 652 | 364 | 388 | 0.170 | 0.093 | 0.144 | 0.020 | 0.033 |
|---|
| TMEM132A |   | 7 | 100 | 33 | 37 | 0.088 | 0.019 | 0.127 | -0.141 | -0.092 |
|---|
| CCNB1 |   | 2 | 391 | 364 | 375 | -0.278 | 0.241 | -0.056 | 0.001 | -0.043 |
|---|
| HOXB2 |   | 1 | 652 | 364 | 388 | -0.062 | 0.024 | -0.143 | 0.076 | -0.184 |
|---|
| BMP2 |   | 7 | 100 | 33 | 37 | -0.155 | -0.011 | 0.097 | 0.148 | -0.069 |
|---|
| CCND1 |   | 12 | 55 | 148 | 150 | 0.122 | -0.258 | 0.074 | 0.117 | 0.003 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| NUP210 |   | 1 | 652 | 364 | 388 | -0.170 | 0.216 | -0.277 | 0.102 | -0.070 |
|---|
| HLX |   | 2 | 391 | 364 | 375 | 0.086 | 0.133 | -0.072 | 0.180 | 0.169 |
|---|
| CTGF |   | 10 | 67 | 57 | 52 | 0.106 | -0.169 | 0.181 | 0.126 | 0.209 |
|---|
| FCGR1A |   | 2 | 391 | 364 | 375 | -0.037 | 0.297 | -0.237 | -0.097 | 0.060 |
|---|
| MYC |   | 24 | 21 | 148 | 137 | -0.056 | -0.101 | -0.169 | 0.188 | -0.134 |
|---|
| PIN1 |   | 17 | 30 | 1 | 13 | 0.135 | 0.194 | 0.040 | -0.040 | -0.048 |
|---|
| GHR |   | 14 | 42 | 148 | 146 | -0.006 | -0.112 | 0.174 | 0.090 | 0.201 |
|---|
| RBM6 |   | 10 | 67 | 148 | 152 | -0.036 | -0.082 | 0.265 | -0.004 | 0.111 |
|---|
| TMBIM6 |   | 1 | 652 | 364 | 388 | 0.191 | 0.058 | -0.029 | -0.051 | -0.034 |
|---|
| BCL2 |   | 11 | 59 | 364 | 350 | 0.067 | -0.077 | -0.035 | 0.128 | 0.043 |
|---|
| ITGAX |   | 1 | 652 | 364 | 388 | 0.131 | 0.087 | -0.191 | 0.039 | -0.160 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
| PDGFRB |   | 3 | 265 | 262 | 270 | 0.009 | -0.094 | 0.224 | 0.127 | 0.189 |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| GREM1 |   | 5 | 148 | 57 | 63 | 0.213 | -0.073 | 0.200 | 0.028 | 0.261 |
|---|
| KDM5B |   | 3 | 265 | 339 | 348 | 0.199 | -0.011 | 0.279 | -0.047 | 0.161 |
|---|
| SKP2 |   | 14 | 42 | 148 | 146 | -0.228 | 0.092 | -0.043 | 0.237 | -0.009 |
|---|
| E2F3 |   | 5 | 148 | 364 | 358 | -0.126 | 0.041 | -0.091 | 0.177 | 0.050 |
|---|
| PTPN12 |   | 2 | 391 | 262 | 283 | 0.048 | -0.159 | 0.153 | 0.031 | 0.042 |
|---|
| TM7SF2 |   | 1 | 652 | 364 | 388 | -0.095 | -0.064 | -0.016 | 0.022 | -0.048 |
|---|
| ATXN1 |   | 6 | 120 | 364 | 355 | 0.171 | 0.026 | 0.192 | 0.175 | 0.163 |
|---|
| UTP3 |   | 1 | 652 | 364 | 388 | 0.166 | 0.090 | 0.098 | 0.164 | -0.172 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| FOS |   | 20 | 24 | 148 | 138 | 0.060 | -0.137 | 0.129 | 0.185 | 0.234 |
|---|
| CDK1 |   | 16 | 34 | 206 | 194 | -0.138 | 0.265 | -0.195 | 0.106 | 0.031 |
|---|
| PBX2 |   | 1 | 652 | 364 | 388 | -0.175 | -0.122 | -0.073 | 0.264 | 0.092 |
|---|
| ANPEP |   | 1 | 652 | 364 | 388 | 0.053 | 0.032 | 0.030 | 0.317 | -0.121 |
|---|
| TPR |   | 10 | 67 | 148 | 152 | 0.118 | -0.130 | 0.120 | -0.036 | 0.156 |
|---|
| FGFR2 |   | 1 | 652 | 364 | 388 | -0.148 | -0.024 | -0.001 | 0.129 | 0.009 |
|---|
| PPP1R15A |   | 1 | 652 | 364 | 388 | 0.086 | 0.170 | -0.001 | -0.018 | 0.081 |
|---|
| CDH5 |   | 2 | 391 | 364 | 375 | 0.009 | -0.159 | 0.023 | 0.180 | 0.033 |
|---|
| ITPR1 |   | 3 | 265 | 182 | 189 | -0.002 | -0.096 | 0.077 | 0.015 | 0.008 |
|---|
| FLT1 |   | 3 | 265 | 364 | 362 | -0.068 | 0.109 | 0.069 | 0.124 | 0.177 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| KLF5 |   | 25 | 19 | 57 | 50 | -0.005 | 0.077 | 0.117 | -0.007 | -0.006 |
|---|
| ITGB4 |   | 6 | 120 | 364 | 355 | 0.044 | 0.021 | 0.313 | -0.134 | -0.114 |
|---|
| CDH2 |   | 7 | 100 | 33 | 37 | -0.228 | 0.169 | 0.109 | -0.016 | 0.155 |
|---|
| CDKN2A |   | 10 | 67 | 364 | 351 | -0.037 | 0.263 | -0.113 | 0.111 | -0.105 |
|---|
| DDIT3 |   | 1 | 652 | 364 | 388 | 0.020 | 0.166 | -0.089 | -0.021 | -0.015 |
|---|
| CTNND2 |   | 1 | 652 | 364 | 388 | -0.124 | 0.007 | 0.078 | -0.046 | -0.291 |
|---|
| BAG1 |   | 1 | 652 | 364 | 388 | -0.115 | 0.168 | 0.025 | 0.160 | 0.130 |
|---|
GO Enrichment output for subnetwork 562 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.355E-09 | 1.462E-05 |
|---|
| interphase | GO:0051325 |  | 7.807E-09 | 8.978E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.406E-08 | 1.078E-05 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 6.36E-08 | 3.657E-05 |
|---|
| regulation of tissue remodeling | GO:0034103 |  | 7.267E-08 | 3.343E-05 |
|---|
| cell cycle arrest | GO:0007050 |  | 1.16E-07 | 4.448E-05 |
|---|
| regulation of ossification | GO:0030278 |  | 6.253E-07 | 2.054E-04 |
|---|
| morphogenesis of a branching structure | GO:0001763 |  | 9.182E-07 | 2.64E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.17E-06 | 2.989E-04 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 1.874E-06 | 4.31E-04 |
|---|
| C21-steroid hormone biosynthetic process | GO:0006700 |  | 1.874E-06 | 3.918E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.651E-09 | 4.033E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.063E-09 | 3.741E-06 |
|---|
| morphogenesis of a branching structure | GO:0001763 |  | 7.465E-09 | 6.079E-06 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 1.445E-08 | 8.824E-06 |
|---|
| regulation of tissue remodeling | GO:0034103 |  | 1.636E-08 | 7.994E-06 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 4.096E-08 | 1.668E-05 |
|---|
| branching morphogenesis of a tube | GO:0048754 |  | 9.71E-08 | 3.389E-05 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 1.219E-07 | 3.724E-05 |
|---|
| regulation of ossification | GO:0030278 |  | 1.856E-07 | 5.037E-05 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 4.573E-07 | 1.117E-04 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 5.722E-07 | 1.271E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.518E-09 | 6.059E-06 |
|---|
| interphase | GO:0051325 |  | 3.326E-09 | 4.002E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.357E-09 | 4.296E-06 |
|---|
| morphogenesis of a branching structure | GO:0001763 |  | 1.303E-08 | 7.837E-06 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 2.517E-08 | 1.211E-05 |
|---|
| regulation of tissue remodeling | GO:0034103 |  | 2.85E-08 | 1.143E-05 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 5.692E-08 | 1.956E-05 |
|---|
| cell cycle arrest | GO:0007050 |  | 7.557E-08 | 2.273E-05 |
|---|
| branching morphogenesis of a tube | GO:0048754 |  | 1.567E-07 | 4.19E-05 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 1.693E-07 | 4.074E-05 |
|---|
| regulation of ossification | GO:0030278 |  | 2.99E-07 | 6.541E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.355E-09 | 1.462E-05 |
|---|
| interphase | GO:0051325 |  | 7.807E-09 | 8.978E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.406E-08 | 1.078E-05 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 6.36E-08 | 3.657E-05 |
|---|
| regulation of tissue remodeling | GO:0034103 |  | 7.267E-08 | 3.343E-05 |
|---|
| cell cycle arrest | GO:0007050 |  | 1.16E-07 | 4.448E-05 |
|---|
| regulation of ossification | GO:0030278 |  | 6.253E-07 | 2.054E-04 |
|---|
| morphogenesis of a branching structure | GO:0001763 |  | 9.182E-07 | 2.64E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.17E-06 | 2.989E-04 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 1.874E-06 | 4.31E-04 |
|---|
| C21-steroid hormone biosynthetic process | GO:0006700 |  | 1.874E-06 | 3.918E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.355E-09 | 1.462E-05 |
|---|
| interphase | GO:0051325 |  | 7.807E-09 | 8.978E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.406E-08 | 1.078E-05 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 6.36E-08 | 3.657E-05 |
|---|
| regulation of tissue remodeling | GO:0034103 |  | 7.267E-08 | 3.343E-05 |
|---|
| cell cycle arrest | GO:0007050 |  | 1.16E-07 | 4.448E-05 |
|---|
| regulation of ossification | GO:0030278 |  | 6.253E-07 | 2.054E-04 |
|---|
| morphogenesis of a branching structure | GO:0001763 |  | 9.182E-07 | 2.64E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.17E-06 | 2.989E-04 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 1.874E-06 | 4.31E-04 |
|---|
| C21-steroid hormone biosynthetic process | GO:0006700 |  | 1.874E-06 | 3.918E-04 |
|---|


