Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 549
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2806 | 1.244e-02 | 8.650e-04 | 3.771e-01 | 4.058e-06 |
|---|
| IPC | 0.4415 | 1.911e-01 | 6.456e-02 | 9.003e-01 | 1.111e-02 |
|---|
| Loi | 0.4301 | 4.180e-04 | 0.000e+00 | 8.086e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6255 | 0.000e+00 | 0.000e+00 | 6.881e-02 | 0.000e+00 |
|---|
| Wang | 0.2337 | 1.860e-02 | 1.151e-01 | 3.415e-01 | 7.311e-04 |
|---|
Expression data for subnetwork 549 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
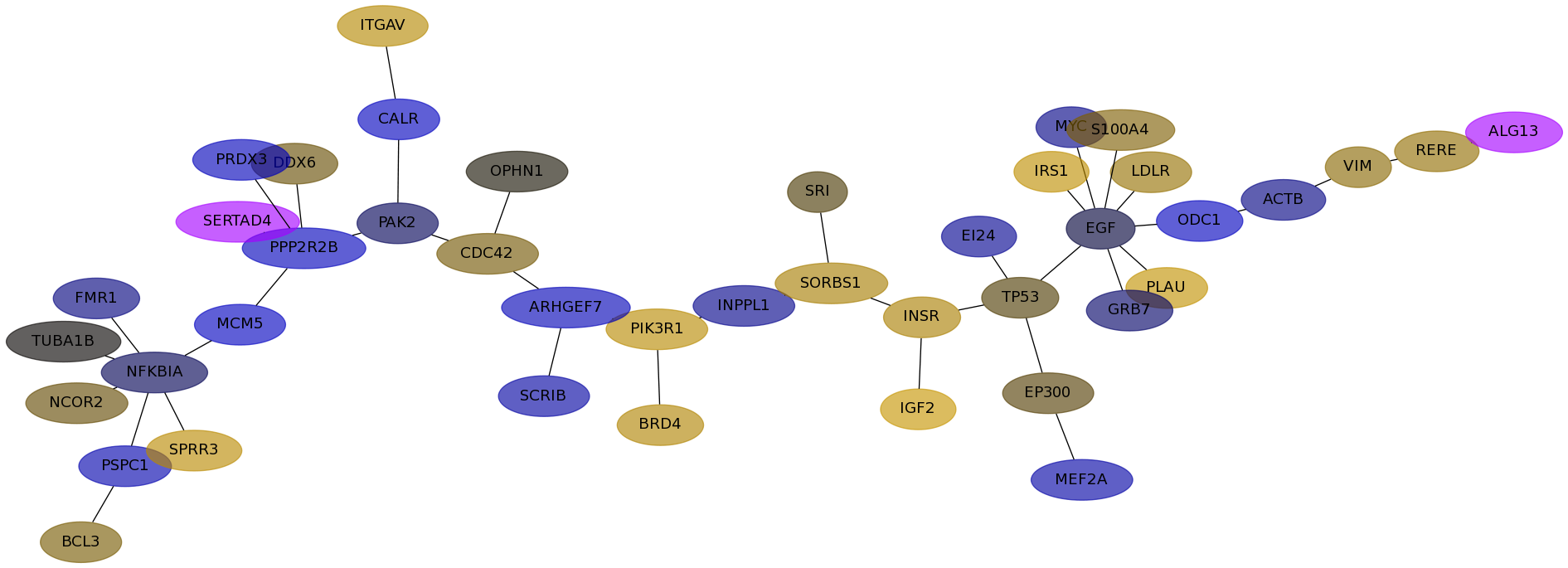
Score for each gene in subnetwork 549 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| NCOR2 |   | 11 | 59 | 283 | 274 | 0.024 | -0.096 | 0.211 | -0.163 | 0.114 |
|---|
| INPPL1 |   | 2 | 391 | 679 | 678 | -0.065 | -0.122 | 0.072 | -0.087 | 0.156 |
|---|
| SORBS1 |   | 17 | 30 | 182 | 178 | 0.124 | -0.043 | 0.015 | 0.314 | 0.200 |
|---|
| ARHGEF7 |   | 12 | 55 | 492 | 475 | -0.180 | -0.074 | -0.061 | 0.208 | 0.015 |
|---|
| EGF |   | 11 | 59 | 398 | 383 | -0.009 | -0.212 | 0.182 | -0.276 | 0.005 |
|---|
| LDLR |   | 3 | 265 | 679 | 667 | 0.085 | -0.041 | 0.218 | 0.039 | 0.064 |
|---|
| OPHN1 |   | 5 | 148 | 339 | 337 | 0.003 | 0.062 | 0.278 | 0.194 | -0.182 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| MEF2A |   | 5 | 148 | 283 | 287 | -0.123 | 0.022 | 0.036 | 0.265 | 0.124 |
|---|
| CALR |   | 10 | 67 | 117 | 121 | -0.216 | -0.105 | 0.002 | -0.061 | -0.075 |
|---|
| SPRR3 |   | 1 | 652 | 679 | 701 | 0.195 | 0.156 | 0.038 | -0.055 | -0.140 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| PLAU |   | 6 | 120 | 444 | 438 | 0.238 | 0.187 | 0.153 | 0.141 | 0.192 |
|---|
| DDX6 |   | 3 | 265 | 283 | 292 | 0.027 | 0.028 | 0.162 | 0.157 | 0.084 |
|---|
| MYC |   | 24 | 21 | 148 | 137 | -0.056 | -0.101 | -0.169 | 0.188 | -0.134 |
|---|
| MCM5 |   | 1 | 652 | 679 | 701 | -0.231 | 0.044 | -0.166 | 0.034 | 0.140 |
|---|
| ALG13 |   | 1 | 652 | 679 | 701 | undef | 0.106 | 0.114 | undef | undef |
|---|
| PAK2 |   | 20 | 24 | 1 | 11 | -0.019 | 0.014 | 0.161 | 0.081 | 0.062 |
|---|
| EI24 |   | 8 | 84 | 339 | 336 | -0.074 | 0.051 | 0.078 | 0.107 | 0.070 |
|---|
| SRI |   | 5 | 148 | 563 | 553 | 0.013 | 0.270 | -0.040 | 0.181 | 0.057 |
|---|
| BCL3 |   | 2 | 391 | 679 | 678 | 0.042 | -0.025 | 0.175 | -0.150 | -0.048 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| FMR1 |   | 3 | 265 | 679 | 667 | -0.047 | 0.137 | -0.104 | 0.352 | 0.113 |
|---|
| BRD4 |   | 18 | 26 | 1 | 12 | 0.156 | -0.119 | 0.095 | -0.075 | -0.044 |
|---|
| SERTAD4 |   | 7 | 100 | 1 | 20 | undef | 0.173 | 0.169 | undef | undef |
|---|
| S100A4 |   | 6 | 120 | 283 | 282 | 0.047 | 0.163 | -0.116 | 0.096 | 0.148 |
|---|
| PSPC1 |   | 2 | 391 | 679 | 678 | -0.148 | 0.203 | -0.289 | 0.298 | -0.107 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| SCRIB |   | 1 | 652 | 679 | 701 | -0.112 | 0.075 | -0.141 | 0.053 | 0.125 |
|---|
| VIM |   | 2 | 391 | 679 | 678 | 0.062 | -0.159 | 0.075 | 0.207 | 0.015 |
|---|
| ODC1 |   | 5 | 148 | 398 | 387 | -0.220 | -0.006 | -0.099 | 0.209 | -0.107 |
|---|
| RERE |   | 4 | 198 | 634 | 615 | 0.077 | -0.284 | 0.209 | -0.027 | 0.074 |
|---|
| TUBA1B |   | 7 | 100 | 182 | 180 | 0.001 | 0.116 | 0.094 | 0.249 | 0.128 |
|---|
| IRS1 |   | 14 | 42 | 1 | 14 | 0.217 | -0.219 | 0.065 | 0.165 | 0.064 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| PRDX3 |   | 2 | 391 | 182 | 195 | -0.194 | 0.038 | -0.059 | 0.173 | -0.143 |
|---|
| GRB7 |   | 13 | 47 | 57 | 51 | -0.028 | 0.021 | 0.115 | -0.072 | 0.129 |
|---|
| NFKBIA |   | 2 | 391 | 679 | 678 | -0.018 | 0.133 | -0.146 | 0.007 | -0.334 |
|---|
| ACTB |   | 7 | 100 | 398 | 385 | -0.052 | -0.039 | 0.283 | -0.061 | 0.003 |
|---|
| ITGAV |   | 2 | 391 | 679 | 678 | 0.174 | -0.051 | 0.203 | 0.155 | 0.105 |
|---|
| IGF2 |   | 6 | 120 | 532 | 514 | 0.261 | -0.022 | 0.172 | 0.145 | 0.116 |
|---|
GO Enrichment output for subnetwork 549 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.112E-09 | 2.559E-06 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.071E-08 | 1.231E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.557E-08 | 1.193E-05 |
|---|
| response to insulin stimulus | GO:0032868 |  | 4.694E-08 | 2.699E-05 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 2.652E-07 | 1.22E-04 |
|---|
| nuclear migration | GO:0007097 |  | 9.88E-07 | 3.787E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 9.88E-07 | 3.246E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.271E-06 | 3.653E-04 |
|---|
| regulation of DNA binding | GO:0051101 |  | 1.608E-06 | 4.11E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.969E-06 | 4.529E-04 |
|---|
| protein import into nucleus. translocation | GO:0000060 |  | 3.084E-06 | 6.448E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 4.9E-10 | 1.197E-06 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 4.427E-09 | 5.407E-06 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.042E-08 | 8.487E-06 |
|---|
| response to insulin stimulus | GO:0032868 |  | 1.678E-08 | 1.025E-05 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 1.115E-07 | 5.448E-05 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 6.122E-07 | 2.493E-04 |
|---|
| regulation of DNA binding | GO:0051101 |  | 8.04E-07 | 2.806E-04 |
|---|
| nuclear migration | GO:0007097 |  | 8.631E-07 | 2.636E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 8.631E-07 | 2.343E-04 |
|---|
| protein import into nucleus. translocation | GO:0000060 |  | 1.797E-06 | 4.391E-04 |
|---|
| maintenance of location | GO:0051235 |  | 2.037E-06 | 4.524E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 5.283E-10 | 1.271E-06 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 5.159E-09 | 6.206E-06 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 6.518E-09 | 5.228E-06 |
|---|
| response to insulin stimulus | GO:0032868 |  | 2.022E-08 | 1.216E-05 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 1.145E-07 | 5.511E-05 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 6.775E-07 | 2.717E-04 |
|---|
| nuclear migration | GO:0007097 |  | 1.02E-06 | 3.507E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.02E-06 | 3.068E-04 |
|---|
| regulation of DNA binding | GO:0051101 |  | 1.03E-06 | 2.753E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.78E-06 | 4.284E-04 |
|---|
| protein import into nucleus. translocation | GO:0000060 |  | 1.884E-06 | 4.121E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.112E-09 | 2.559E-06 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.071E-08 | 1.231E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.557E-08 | 1.193E-05 |
|---|
| response to insulin stimulus | GO:0032868 |  | 4.694E-08 | 2.699E-05 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 2.652E-07 | 1.22E-04 |
|---|
| nuclear migration | GO:0007097 |  | 9.88E-07 | 3.787E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 9.88E-07 | 3.246E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.271E-06 | 3.653E-04 |
|---|
| regulation of DNA binding | GO:0051101 |  | 1.608E-06 | 4.11E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.969E-06 | 4.529E-04 |
|---|
| protein import into nucleus. translocation | GO:0000060 |  | 3.084E-06 | 6.448E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.112E-09 | 2.559E-06 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.071E-08 | 1.231E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.557E-08 | 1.193E-05 |
|---|
| response to insulin stimulus | GO:0032868 |  | 4.694E-08 | 2.699E-05 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 2.652E-07 | 1.22E-04 |
|---|
| nuclear migration | GO:0007097 |  | 9.88E-07 | 3.787E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 9.88E-07 | 3.246E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.271E-06 | 3.653E-04 |
|---|
| regulation of DNA binding | GO:0051101 |  | 1.608E-06 | 4.11E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.969E-06 | 4.529E-04 |
|---|
| protein import into nucleus. translocation | GO:0000060 |  | 3.084E-06 | 6.448E-04 |
|---|


