Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 517
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2826 | 1.156e-02 | 7.650e-04 | 3.672e-01 | 3.247e-06 |
|---|
| IPC | 0.4444 | 1.871e-01 | 6.233e-02 | 8.993e-01 | 1.049e-02 |
|---|
| Loi | 0.4210 | 6.010e-04 | 0.000e+00 | 8.739e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6140 | 0.000e+00 | 0.000e+00 | 8.665e-02 | 0.000e+00 |
|---|
| Wang | 0.2305 | 2.078e-02 | 1.232e-01 | 3.580e-01 | 9.160e-04 |
|---|
Expression data for subnetwork 517 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
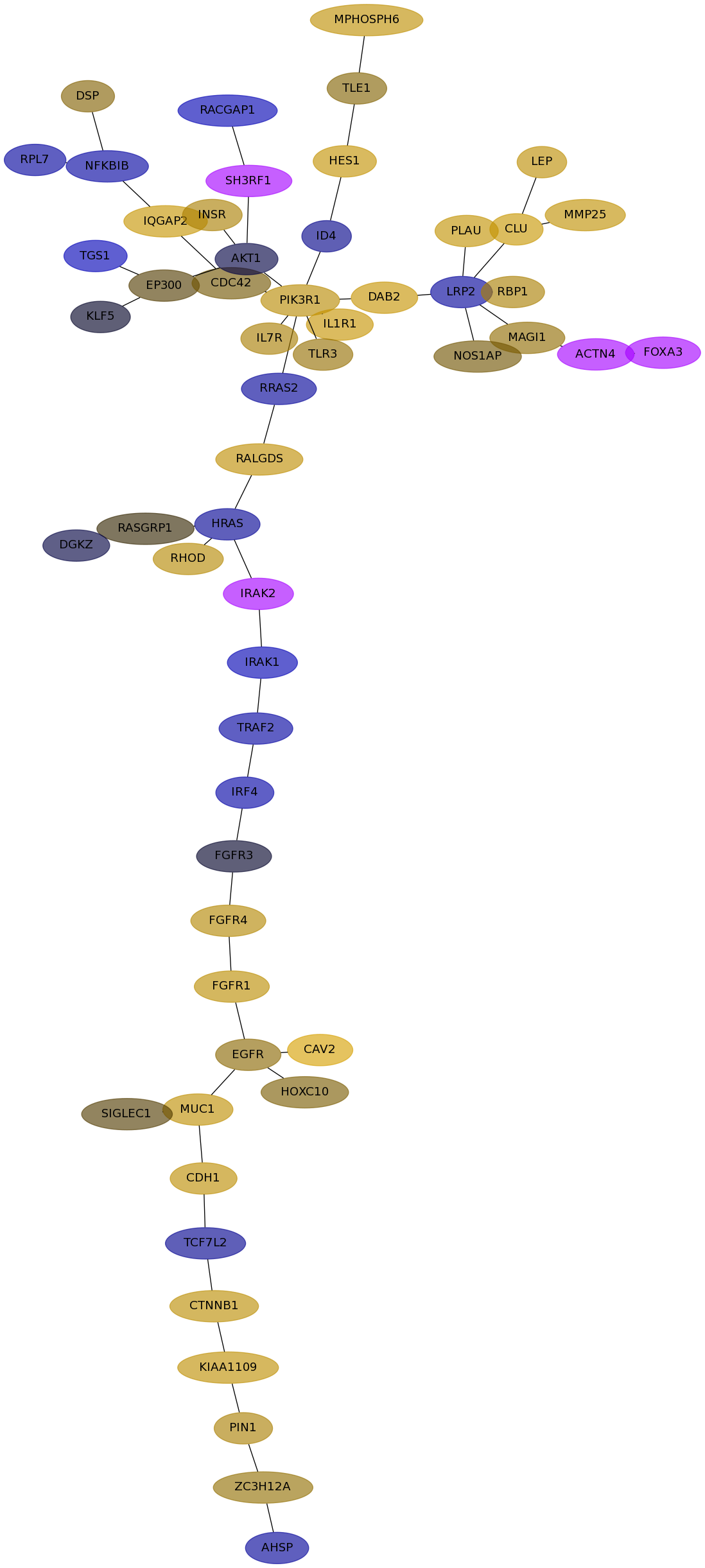
Score for each gene in subnetwork 517 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| DGKZ |   | 1 | 652 | 1177 | 1191 | -0.011 | 0.093 | -0.043 | -0.081 | -0.064 |
|---|
| RHOD |   | 1 | 652 | 1177 | 1191 | 0.174 | 0.172 | 0.036 | 0.099 | 0.116 |
|---|
| TRAF2 |   | 6 | 120 | 206 | 200 | -0.107 | 0.185 | -0.172 | -0.013 | -0.092 |
|---|
| FOXA3 |   | 6 | 120 | 657 | 631 | undef | 0.192 | 0.289 | undef | undef |
|---|
| LRP2 |   | 2 | 391 | 1177 | 1166 | -0.092 | 0.193 | -0.042 | 0.001 | 0.125 |
|---|
| RALGDS |   | 5 | 148 | 647 | 621 | 0.212 | -0.011 | 0.199 | 0.033 | -0.009 |
|---|
| RACGAP1 |   | 3 | 265 | 778 | 770 | -0.166 | 0.283 | -0.164 | 0.073 | 0.188 |
|---|
| ID4 |   | 13 | 47 | 444 | 430 | -0.060 | -0.179 | 0.137 | 0.241 | 0.036 |
|---|
| AHSP |   | 1 | 652 | 1177 | 1191 | -0.090 | 0.009 | -0.131 | -0.125 | undef |
|---|
| NOS1AP |   | 1 | 652 | 1177 | 1191 | 0.035 | 0.041 | 0.050 | -0.058 | -0.080 |
|---|
| IQGAP2 |   | 16 | 34 | 117 | 119 | 0.264 | -0.170 | -0.051 | -0.112 | 0.049 |
|---|
| FGFR3 |   | 5 | 148 | 963 | 938 | -0.006 | 0.160 | 0.071 | 0.018 | -0.111 |
|---|
| IRF4 |   | 2 | 391 | 1094 | 1081 | -0.123 | 0.153 | -0.250 | -0.172 | -0.103 |
|---|
| DAB2 |   | 13 | 47 | 1 | 15 | 0.258 | -0.175 | 0.067 | -0.022 | 0.071 |
|---|
| TLR3 |   | 2 | 391 | 206 | 220 | 0.081 | 0.196 | -0.111 | -0.089 | -0.103 |
|---|
| IRAK1 |   | 4 | 198 | 206 | 205 | -0.170 | 0.266 | -0.023 | 0.092 | -0.102 |
|---|
| PLAU |   | 6 | 120 | 444 | 438 | 0.238 | 0.187 | 0.153 | 0.141 | 0.192 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| PIN1 |   | 17 | 30 | 1 | 13 | 0.135 | 0.194 | 0.040 | -0.040 | -0.048 |
|---|
| LEP |   | 2 | 391 | 117 | 135 | 0.212 | -0.014 | -0.019 | 0.145 | 0.059 |
|---|
| RRAS2 |   | 6 | 120 | 563 | 549 | -0.089 | 0.155 | 0.076 | 0.289 | 0.031 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| MPHOSPH6 |   | 8 | 84 | 756 | 726 | 0.217 | -0.084 | 0.175 | -0.269 | 0.146 |
|---|
| HOXC10 |   | 4 | 198 | 444 | 444 | 0.044 | -0.015 | 0.116 | -0.133 | -0.017 |
|---|
| RASGRP1 |   | 2 | 391 | 1094 | 1081 | 0.008 | -0.076 | -0.084 | 0.066 | -0.109 |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| HES1 |   | 4 | 198 | 746 | 725 | 0.247 | 0.115 | 0.157 | 0.010 | -0.106 |
|---|
| AKT1 |   | 13 | 47 | 444 | 430 | -0.012 | 0.045 | 0.126 | -0.099 | -0.168 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| RPL7 |   | 1 | 652 | 1177 | 1191 | -0.100 | 0.189 | -0.121 | 0.166 | -0.003 |
|---|
| KIAA1109 |   | 2 | 391 | 1106 | 1100 | 0.220 | -0.036 | 0.201 | 0.065 | 0.173 |
|---|
| DSP |   | 7 | 100 | 206 | 198 | 0.052 | 0.054 | 0.235 | -0.119 | 0.053 |
|---|
| TCF7L2 |   | 17 | 30 | 271 | 264 | -0.071 | -0.021 | 0.143 | 0.280 | 0.101 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| IL1R1 |   | 8 | 84 | 536 | 515 | 0.257 | -0.211 | 0.178 | -0.071 | 0.003 |
|---|
| ZC3H12A |   | 2 | 391 | 1052 | 1044 | 0.079 | 0.034 | 0.001 | 0.094 | -0.114 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| IL7R |   | 14 | 42 | 339 | 334 | 0.130 | -0.151 | -0.112 | -0.130 | -0.160 |
|---|
| SH3RF1 |   | 14 | 42 | 117 | 120 | undef | 0.144 | 0.238 | undef | undef |
|---|
| IRAK2 |   | 3 | 265 | 1094 | 1070 | undef | 0.183 | 0.023 | undef | undef |
|---|
| TLE1 |   | 2 | 391 | 1177 | 1166 | 0.054 | 0.183 | 0.050 | 0.030 | 0.055 |
|---|
| ACTN4 |   | 6 | 120 | 859 | 834 | undef | 0.035 | 0.180 | undef | undef |
|---|
| MMP25 |   | 1 | 652 | 1177 | 1191 | 0.225 | -0.006 | -0.061 | 0.169 | 0.038 |
|---|
| KLF5 |   | 25 | 19 | 57 | 50 | -0.005 | 0.077 | 0.117 | -0.007 | -0.006 |
|---|
| CLU |   | 10 | 67 | 117 | 121 | 0.280 | -0.208 | 0.119 | -0.153 | -0.234 |
|---|
| NFKBIB |   | 1 | 652 | 1177 | 1191 | -0.107 | -0.008 | -0.192 | -0.185 | -0.239 |
|---|
| MAGI1 |   | 2 | 391 | 937 | 928 | 0.073 | 0.094 | 0.020 | 0.073 | 0.004 |
|---|
| TGS1 |   | 1 | 652 | 1177 | 1191 | -0.178 | -0.060 | -0.044 | 0.232 | 0.269 |
|---|
| HRAS |   | 3 | 265 | 1094 | 1070 | -0.081 | 0.037 | 0.090 | 0.098 | -0.005 |
|---|
| SIGLEC1 |   | 3 | 265 | 963 | 950 | 0.017 | 0.045 | -0.114 | -0.107 | -0.052 |
|---|
| RBP1 |   | 2 | 391 | 1177 | 1166 | 0.133 | -0.002 | -0.005 | 0.112 | -0.089 |
|---|
GO Enrichment output for subnetwork 517 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 8.018E-08 | 1.844E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.159E-07 | 2.483E-04 |
|---|
| activation of NF-kappaB-inducing kinase activity | GO:0007250 |  | 9.655E-07 | 7.402E-04 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 1.74E-06 | 1.001E-03 |
|---|
| nuclear migration | GO:0007097 |  | 1.801E-06 | 8.285E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.801E-06 | 6.904E-04 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 2.265E-06 | 7.444E-04 |
|---|
| cell growth | GO:0016049 |  | 2.732E-06 | 7.855E-04 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 2.9E-06 | 7.411E-04 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 3.587E-06 | 8.251E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 3.587E-06 | 7.501E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 7.602E-10 | 1.857E-06 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 3.229E-08 | 3.945E-05 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.831E-07 | 1.491E-04 |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 2.671E-07 | 1.632E-04 |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 8.562E-07 | 4.184E-04 |
|---|
| activation of NF-kappaB-inducing kinase activity | GO:0007250 |  | 8.769E-07 | 3.57E-04 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.124E-06 | 3.921E-04 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 1.74E-06 | 5.312E-04 |
|---|
| nuclear migration | GO:0007097 |  | 1.74E-06 | 4.722E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.74E-06 | 4.25E-04 |
|---|
| pancreas development | GO:0031016 |  | 1.767E-06 | 3.923E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 4.478E-10 | 1.077E-06 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.656E-08 | 3.195E-05 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 8.937E-08 | 7.168E-05 |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 2.838E-07 | 1.707E-04 |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 8.052E-07 | 3.875E-04 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 9.314E-07 | 3.735E-04 |
|---|
| activation of NF-kappaB-inducing kinase activity | GO:0007250 |  | 9.314E-07 | 3.201E-04 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 1.823E-06 | 5.484E-04 |
|---|
| nuclear migration | GO:0007097 |  | 1.823E-06 | 4.874E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.823E-06 | 4.387E-04 |
|---|
| pancreas development | GO:0031016 |  | 1.876E-06 | 4.104E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 8.018E-08 | 1.844E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.159E-07 | 2.483E-04 |
|---|
| activation of NF-kappaB-inducing kinase activity | GO:0007250 |  | 9.655E-07 | 7.402E-04 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 1.74E-06 | 1.001E-03 |
|---|
| nuclear migration | GO:0007097 |  | 1.801E-06 | 8.285E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.801E-06 | 6.904E-04 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 2.265E-06 | 7.444E-04 |
|---|
| cell growth | GO:0016049 |  | 2.732E-06 | 7.855E-04 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 2.9E-06 | 7.411E-04 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 3.587E-06 | 8.251E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 3.587E-06 | 7.501E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 8.018E-08 | 1.844E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.159E-07 | 2.483E-04 |
|---|
| activation of NF-kappaB-inducing kinase activity | GO:0007250 |  | 9.655E-07 | 7.402E-04 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 1.74E-06 | 1.001E-03 |
|---|
| nuclear migration | GO:0007097 |  | 1.801E-06 | 8.285E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.801E-06 | 6.904E-04 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 2.265E-06 | 7.444E-04 |
|---|
| cell growth | GO:0016049 |  | 2.732E-06 | 7.855E-04 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 2.9E-06 | 7.411E-04 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 3.587E-06 | 8.251E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 3.587E-06 | 7.501E-04 |
|---|


