Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 502
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2852 | 1.056e-02 | 6.590e-04 | 3.554e-01 | 2.474e-06 |
|---|
| IPC | 0.4418 | 1.907e-01 | 6.434e-02 | 9.002e-01 | 1.105e-02 |
|---|
| Loi | 0.4180 | 6.770e-04 | 0.000e+00 | 8.965e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6082 | 1.000e-06 | 0.000e+00 | 9.690e-02 | 0.000e+00 |
|---|
| Wang | 0.2287 | 2.205e-02 | 1.277e-01 | 3.670e-01 | 1.034e-03 |
|---|
Expression data for subnetwork 502 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
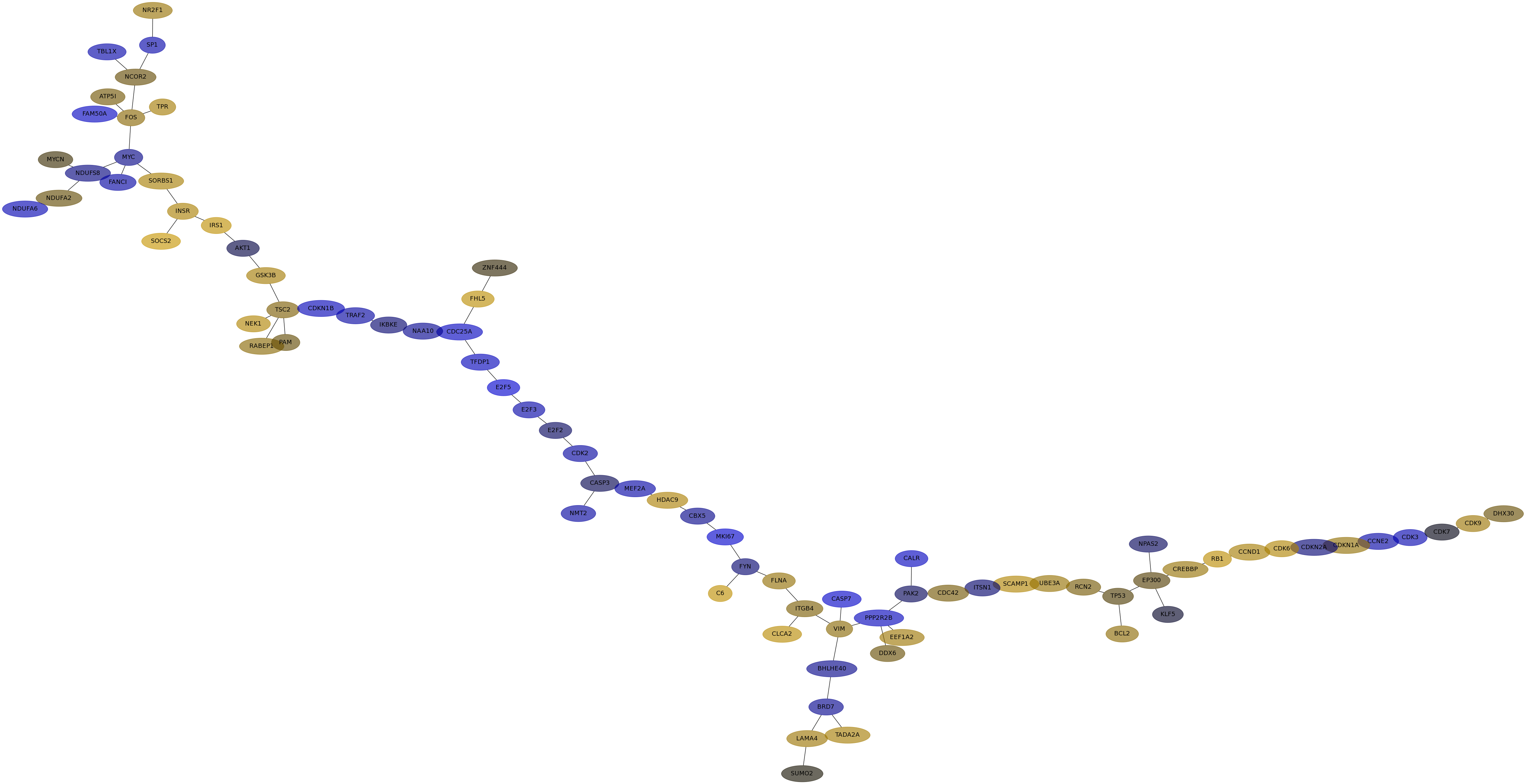
Score for each gene in subnetwork 502 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| CCNE2 |   | 8 | 84 | 536 | 515 | -0.123 | 0.136 | -0.178 | 0.241 | -0.055 |
|---|
| CASP3 |   | 4 | 198 | 129 | 130 | -0.015 | 0.234 | -0.032 | -0.142 | -0.084 |
|---|
| SORBS1 |   | 17 | 30 | 182 | 178 | 0.124 | -0.043 | 0.015 | 0.314 | 0.200 |
|---|
| TSC2 |   | 3 | 265 | 57 | 71 | 0.046 | 0.236 | 0.044 | 0.114 | -0.102 |
|---|
| SCAMP1 |   | 1 | 652 | 1338 | 1339 | 0.157 | 0.240 | 0.146 | 0.216 | 0.065 |
|---|
| NPAS2 |   | 3 | 265 | 364 | 362 | -0.019 | 0.116 | 0.253 | -0.157 | -0.093 |
|---|
| E2F5 |   | 1 | 652 | 1338 | 1339 | -0.315 | 0.048 | -0.087 | 0.002 | -0.074 |
|---|
| SUMO2 |   | 12 | 55 | 148 | 150 | 0.003 | 0.117 | 0.062 | 0.294 | -0.096 |
|---|
| SOCS2 |   | 4 | 198 | 859 | 837 | 0.262 | -0.130 | -0.028 | 0.051 | 0.027 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| ATP5I |   | 1 | 652 | 1338 | 1339 | 0.035 | 0.204 | -0.002 | 0.064 | 0.001 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| CDK3 |   | 5 | 148 | 129 | 129 | -0.151 | -0.076 | 0.189 | -0.041 | 0.082 |
|---|
| FANCI |   | 4 | 198 | 832 | 809 | -0.111 | 0.145 | -0.194 | 0.104 | -0.036 |
|---|
| CCND1 |   | 12 | 55 | 148 | 150 | 0.122 | -0.258 | 0.074 | 0.117 | 0.003 |
|---|
| CREBBP |   | 5 | 148 | 444 | 440 | 0.084 | -0.104 | 0.102 | 0.093 | 0.002 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| CDKN1B |   | 2 | 391 | 937 | 928 | -0.180 | 0.120 | -0.171 | 0.088 | -0.005 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| NDUFA6 |   | 1 | 652 | 1338 | 1339 | -0.163 | 0.100 | 0.003 | -0.060 | 0.177 |
|---|
| DHX30 |   | 1 | 652 | 1338 | 1339 | 0.027 | -0.213 | 0.084 | 0.253 | 0.096 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
| TADA2A |   | 1 | 652 | 1338 | 1339 | 0.122 | 0.135 | -0.054 | 0.204 | -0.127 |
|---|
| ITSN1 |   | 1 | 652 | 1338 | 1339 | -0.030 | -0.044 | 0.026 | 0.113 | 0.124 |
|---|
| ZNF444 |   | 1 | 652 | 1338 | 1339 | 0.007 | 0.055 | 0.151 | -0.054 | -0.160 |
|---|
| LAMA4 |   | 3 | 265 | 563 | 556 | 0.094 | -0.016 | 0.138 | 0.209 | 0.343 |
|---|
| AKT1 |   | 13 | 47 | 444 | 430 | -0.012 | 0.045 | 0.126 | -0.099 | -0.168 |
|---|
| MKI67 |   | 3 | 265 | 778 | 770 | -0.295 | 0.205 | -0.123 | 0.176 | 0.018 |
|---|
| E2F3 |   | 5 | 148 | 364 | 358 | -0.126 | 0.041 | -0.091 | 0.177 | 0.050 |
|---|
| VIM |   | 2 | 391 | 679 | 678 | 0.062 | -0.159 | 0.075 | 0.207 | 0.015 |
|---|
| CDK7 |   | 4 | 198 | 129 | 130 | -0.002 | 0.105 | 0.184 | 0.017 | -0.097 |
|---|
| FLNA |   | 7 | 100 | 206 | 198 | 0.077 | 0.081 | 0.107 | 0.148 | 0.126 |
|---|
| RCN2 |   | 6 | 120 | 756 | 731 | 0.037 | 0.059 | 0.051 | 0.176 | -0.098 |
|---|
| FYN |   | 3 | 265 | 182 | 189 | -0.032 | -0.171 | -0.167 | 0.073 | 0.110 |
|---|
| CLCA2 |   | 5 | 148 | 206 | 202 | 0.188 | -0.176 | 0.243 | -0.153 | -0.000 |
|---|
| PAM |   | 2 | 391 | 1065 | 1059 | 0.024 | -0.035 | 0.307 | 0.200 | 0.104 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| NAA10 |   | 1 | 652 | 1338 | 1339 | -0.072 | 0.191 | -0.048 | 0.013 | -0.095 |
|---|
| KLF5 |   | 25 | 19 | 57 | 50 | -0.005 | 0.077 | 0.117 | -0.007 | -0.006 |
|---|
| E2F2 |   | 4 | 198 | 129 | 130 | -0.021 | 0.212 | -0.047 | 0.162 | -0.092 |
|---|
| NCOR2 |   | 11 | 59 | 283 | 274 | 0.024 | -0.096 | 0.211 | -0.163 | 0.114 |
|---|
| TRAF2 |   | 6 | 120 | 206 | 200 | -0.107 | 0.185 | -0.172 | -0.013 | -0.092 |
|---|
| C6 |   | 1 | 652 | 1338 | 1339 | 0.217 | 0.128 | 0.055 | 0.169 | 0.054 |
|---|
| MEF2A |   | 5 | 148 | 283 | 287 | -0.123 | 0.022 | 0.036 | 0.265 | 0.124 |
|---|
| BHLHE40 |   | 1 | 652 | 1338 | 1339 | -0.060 | 0.024 | 0.133 | 0.051 | 0.033 |
|---|
| HDAC9 |   | 1 | 652 | 1338 | 1339 | 0.138 | 0.155 | -0.084 | -0.080 | -0.015 |
|---|
| CALR |   | 10 | 67 | 117 | 121 | -0.216 | -0.105 | 0.002 | -0.061 | -0.075 |
|---|
| FAM50A |   | 1 | 652 | 1338 | 1339 | -0.229 | 0.173 | 0.079 | -0.043 | 0.039 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| UBE3A |   | 2 | 391 | 820 | 815 | 0.085 | -0.043 | -0.018 | 0.183 | 0.017 |
|---|
| NDUFA2 |   | 1 | 652 | 1338 | 1339 | 0.024 | 0.159 | -0.014 | 0.084 | -0.022 |
|---|
| DDX6 |   | 3 | 265 | 283 | 292 | 0.027 | 0.028 | 0.162 | 0.157 | 0.084 |
|---|
| MYC |   | 24 | 21 | 148 | 137 | -0.056 | -0.101 | -0.169 | 0.188 | -0.134 |
|---|
| EEF1A2 |   | 9 | 77 | 182 | 179 | 0.098 | 0.032 | 0.220 | -0.119 | 0.102 |
|---|
| PAK2 |   | 20 | 24 | 1 | 11 | -0.019 | 0.014 | 0.161 | 0.081 | 0.062 |
|---|
| BRD7 |   | 4 | 198 | 906 | 894 | -0.068 | -0.090 | 0.019 | -0.188 | -0.086 |
|---|
| NDUFS8 |   | 1 | 652 | 1338 | 1339 | -0.047 | 0.013 | -0.088 | -0.150 | 0.030 |
|---|
| BCL2 |   | 11 | 59 | 364 | 350 | 0.067 | -0.077 | -0.035 | 0.128 | 0.043 |
|---|
| MYCN |   | 3 | 265 | 859 | 843 | 0.009 | 0.125 | 0.057 | -0.003 | -0.084 |
|---|
| SP1 |   | 16 | 34 | 283 | 272 | -0.129 | 0.222 | -0.254 | 0.239 | 0.082 |
|---|
| CDC25A |   | 5 | 148 | 444 | 440 | -0.220 | 0.113 | -0.170 | 0.151 | -0.003 |
|---|
| FOS |   | 20 | 24 | 148 | 138 | 0.060 | -0.137 | 0.129 | 0.185 | 0.234 |
|---|
| NR2F1 |   | 6 | 120 | 444 | 438 | 0.086 | -0.252 | 0.113 | 0.133 | 0.207 |
|---|
| NMT2 |   | 1 | 652 | 1338 | 1339 | -0.104 | 0.077 | -0.296 | 0.134 | 0.217 |
|---|
| IRS1 |   | 14 | 42 | 1 | 14 | 0.217 | -0.219 | 0.065 | 0.165 | 0.064 |
|---|
| TPR |   | 10 | 67 | 148 | 152 | 0.118 | -0.130 | 0.120 | -0.036 | 0.156 |
|---|
| NEK1 |   | 4 | 198 | 57 | 68 | 0.155 | 0.032 | 0.184 | 0.049 | 0.135 |
|---|
| CASP7 |   | 8 | 84 | 283 | 278 | -0.270 | 0.216 | 0.095 | 0.255 | 0.035 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| IKBKE |   | 8 | 84 | 756 | 726 | -0.033 | 0.227 | -0.041 | -0.095 | 0.079 |
|---|
| CDK9 |   | 4 | 198 | 129 | 130 | 0.095 | -0.120 | 0.179 | 0.182 | -0.135 |
|---|
| TFDP1 |   | 4 | 198 | 339 | 341 | -0.194 | 0.123 | -0.178 | 0.129 | -0.065 |
|---|
| TBL1X |   | 2 | 391 | 1008 | 999 | -0.131 | -0.044 | 0.017 | -0.192 | -0.009 |
|---|
| ITGB4 |   | 6 | 120 | 364 | 355 | 0.044 | 0.021 | 0.313 | -0.134 | -0.114 |
|---|
| CDK2 |   | 16 | 34 | 129 | 124 | -0.104 | 0.196 | -0.096 | 0.109 | 0.362 |
|---|
| CBX5 |   | 2 | 391 | 1292 | 1279 | -0.061 | 0.155 | 0.016 | 0.148 | -0.009 |
|---|
| RABEP1 |   | 1 | 652 | 1338 | 1339 | 0.062 | -0.004 | 0.239 | 0.060 | 0.008 |
|---|
| CDKN2A |   | 10 | 67 | 364 | 351 | -0.037 | 0.263 | -0.113 | 0.111 | -0.105 |
|---|
| FHL5 |   | 1 | 652 | 1338 | 1339 | 0.204 | 0.022 | 0.031 | 0.145 | 0.153 |
|---|
GO Enrichment output for subnetwork 502 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.564E-11 | 1.28E-07 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 2.201E-10 | 2.532E-07 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.746E-09 | 1.338E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.704E-09 | 4.43E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.149E-08 | 5.286E-06 |
|---|
| interphase | GO:0051325 |  | 1.44E-08 | 5.522E-06 |
|---|
| peptide or protein amino-terminal blocking | GO:0018409 |  | 2.046E-08 | 6.721E-06 |
|---|
| N-terminal protein amino acid modification | GO:0031365 |  | 1.02E-07 | 2.933E-05 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 1.306E-07 | 3.337E-05 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.574E-07 | 3.621E-05 |
|---|
| negative regulation of cell size | GO:0045792 |  | 1.975E-07 | 4.13E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.501E-12 | 8.552E-09 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 3.128E-11 | 3.821E-08 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.745E-10 | 4.678E-07 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.987E-10 | 3.657E-07 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 9.012E-10 | 4.403E-07 |
|---|
| peptide or protein amino-terminal blocking | GO:0018409 |  | 4.862E-09 | 1.98E-06 |
|---|
| negative regulation of cell size | GO:0045792 |  | 1.62E-08 | 5.654E-06 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.074E-08 | 6.332E-06 |
|---|
| N-terminal protein amino acid modification | GO:0031365 |  | 3.094E-08 | 8.398E-06 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 5.888E-08 | 1.438E-05 |
|---|
| response to UV | GO:0009411 |  | 6.756E-08 | 1.5E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.186E-12 | 1.969E-08 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 5.748E-11 | 6.915E-08 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 3.505E-10 | 2.811E-07 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.137E-09 | 6.837E-07 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.517E-09 | 7.299E-07 |
|---|
| interphase | GO:0051325 |  | 2.064E-09 | 8.277E-07 |
|---|
| peptide or protein amino-terminal blocking | GO:0018409 |  | 7.534E-09 | 2.59E-06 |
|---|
| negative regulation of cell size | GO:0045792 |  | 3.169E-08 | 9.53E-06 |
|---|
| N-terminal protein amino acid modification | GO:0031365 |  | 3.209E-08 | 8.579E-06 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 3.209E-08 | 7.721E-06 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 8.373E-08 | 1.831E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.564E-11 | 1.28E-07 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 2.201E-10 | 2.532E-07 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.746E-09 | 1.338E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.704E-09 | 4.43E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.149E-08 | 5.286E-06 |
|---|
| interphase | GO:0051325 |  | 1.44E-08 | 5.522E-06 |
|---|
| peptide or protein amino-terminal blocking | GO:0018409 |  | 2.046E-08 | 6.721E-06 |
|---|
| N-terminal protein amino acid modification | GO:0031365 |  | 1.02E-07 | 2.933E-05 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 1.306E-07 | 3.337E-05 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.574E-07 | 3.621E-05 |
|---|
| negative regulation of cell size | GO:0045792 |  | 1.975E-07 | 4.13E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.564E-11 | 1.28E-07 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 2.201E-10 | 2.532E-07 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.746E-09 | 1.338E-06 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.704E-09 | 4.43E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.149E-08 | 5.286E-06 |
|---|
| interphase | GO:0051325 |  | 1.44E-08 | 5.522E-06 |
|---|
| peptide or protein amino-terminal blocking | GO:0018409 |  | 2.046E-08 | 6.721E-06 |
|---|
| N-terminal protein amino acid modification | GO:0031365 |  | 1.02E-07 | 2.933E-05 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 1.306E-07 | 3.337E-05 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.574E-07 | 3.621E-05 |
|---|
| negative regulation of cell size | GO:0045792 |  | 1.975E-07 | 4.13E-05 |
|---|


