Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 427
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2910 | 8.532e-03 | 4.620e-04 | 3.285e-01 | 1.295e-06 |
|---|
| IPC | 0.4151 | 2.295e-01 | 8.777e-02 | 9.095e-01 | 1.832e-02 |
|---|
| Loi | 0.4142 | 7.850e-04 | 0.000e+00 | 9.261e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5808 | 3.000e-06 | 0.000e+00 | 1.571e-01 | 0.000e+00 |
|---|
| Wang | 0.2082 | 4.218e-02 | 1.893e-01 | 4.769e-01 | 3.808e-03 |
|---|
Expression data for subnetwork 427 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
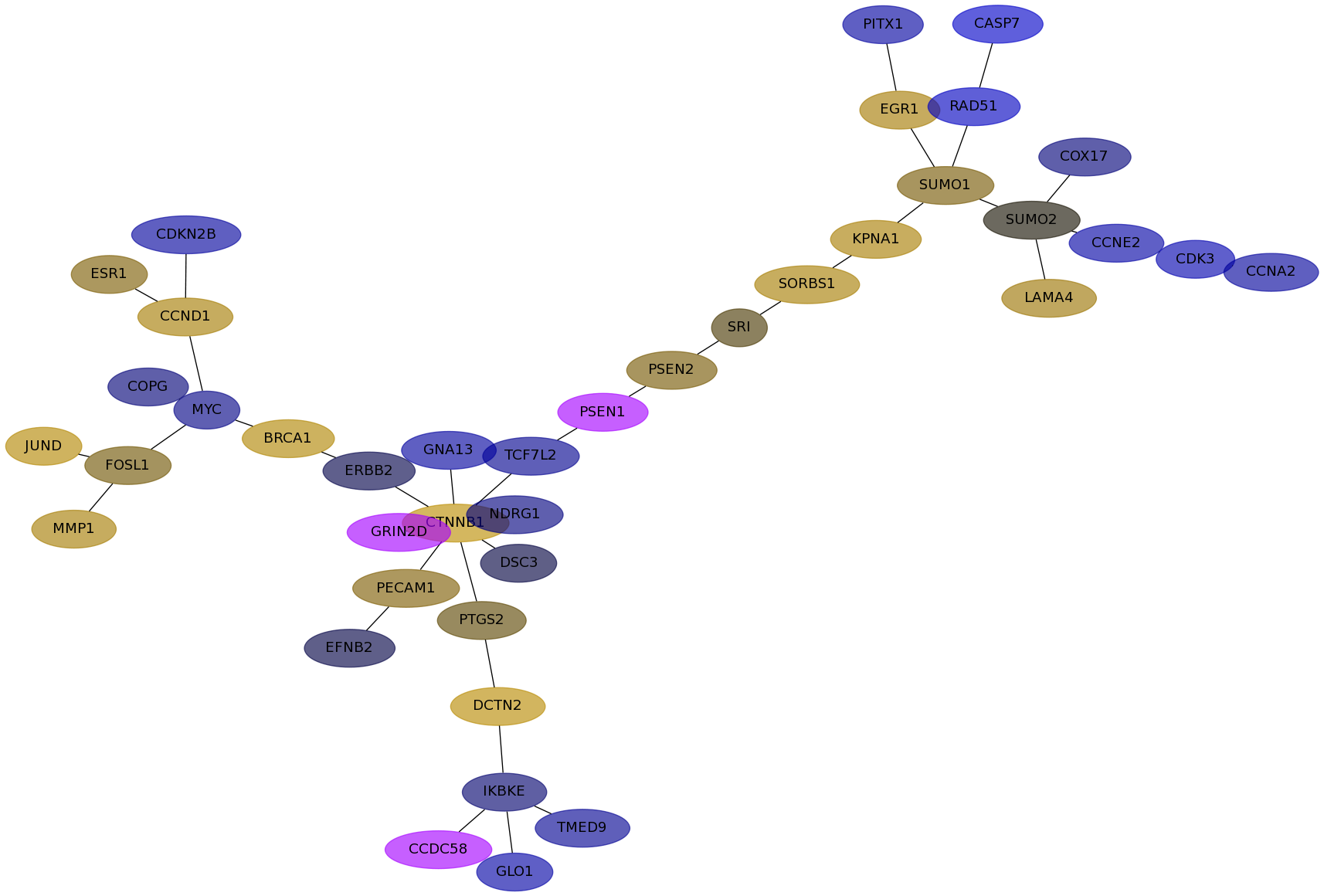
Score for each gene in subnetwork 427 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| CDKN2B |   | 4 | 198 | 937 | 910 | -0.099 | 0.066 | 0.050 | -0.028 | 0.153 |
|---|
| EFNB2 |   | 1 | 652 | 1472 | 1472 | -0.012 | -0.070 | 0.156 | 0.193 | -0.116 |
|---|
| CCNE2 |   | 8 | 84 | 536 | 515 | -0.123 | 0.136 | -0.178 | 0.241 | -0.055 |
|---|
| SORBS1 |   | 17 | 30 | 182 | 178 | 0.124 | -0.043 | 0.015 | 0.314 | 0.200 |
|---|
| EGR1 |   | 11 | 59 | 283 | 274 | 0.118 | -0.184 | 0.259 | 0.130 | 0.122 |
|---|
| CCDC58 |   | 2 | 391 | 1198 | 1184 | undef | 0.241 | -0.030 | undef | undef |
|---|
| NDRG1 |   | 3 | 265 | 756 | 748 | -0.049 | 0.222 | 0.143 | -0.040 | 0.242 |
|---|
| DCTN2 |   | 3 | 265 | 859 | 843 | 0.192 | 0.014 | 0.150 | 0.061 | 0.303 |
|---|
| SUMO2 |   | 12 | 55 | 148 | 150 | 0.003 | 0.117 | 0.062 | 0.294 | -0.096 |
|---|
| ERBB2 |   | 2 | 391 | 715 | 711 | -0.014 | -0.103 | 0.092 | -0.090 | 0.046 |
|---|
| PECAM1 |   | 3 | 265 | 552 | 550 | 0.046 | 0.014 | -0.161 | 0.040 | -0.106 |
|---|
| PSEN1 |   | 5 | 148 | 847 | 821 | undef | -0.041 | 0.110 | undef | undef |
|---|
| CDK3 |   | 5 | 148 | 129 | 129 | -0.151 | -0.076 | 0.189 | -0.041 | 0.082 |
|---|
| SUMO1 |   | 3 | 265 | 1106 | 1083 | 0.038 | 0.245 | -0.007 | 0.144 | 0.118 |
|---|
| JUND |   | 1 | 652 | 1472 | 1472 | 0.171 | -0.077 | 0.091 | -0.075 | -0.005 |
|---|
| CCND1 |   | 12 | 55 | 148 | 150 | 0.122 | -0.258 | 0.074 | 0.117 | 0.003 |
|---|
| CCNA2 |   | 15 | 40 | 148 | 144 | -0.097 | 0.176 | -0.053 | 0.188 | 0.246 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| MYC |   | 24 | 21 | 148 | 137 | -0.056 | -0.101 | -0.169 | 0.188 | -0.134 |
|---|
| SRI |   | 5 | 148 | 563 | 553 | 0.013 | 0.270 | -0.040 | 0.181 | 0.057 |
|---|
| PSEN2 |   | 2 | 391 | 1198 | 1184 | 0.039 | 0.239 | -0.068 | -0.157 | 0.045 |
|---|
| TMED9 |   | 2 | 391 | 1198 | 1184 | -0.076 | 0.142 | 0.057 | -0.078 | 0.321 |
|---|
| DSC3 |   | 2 | 391 | 1198 | 1184 | -0.010 | -0.023 | 0.014 | 0.324 | 0.147 |
|---|
| LAMA4 |   | 3 | 265 | 563 | 556 | 0.094 | -0.016 | 0.138 | 0.209 | 0.343 |
|---|
| MMP1 |   | 3 | 265 | 1039 | 1013 | 0.122 | 0.192 | 0.014 | 0.036 | 0.260 |
|---|
| GLO1 |   | 3 | 265 | 859 | 843 | -0.123 | 0.266 | -0.089 | 0.135 | 0.076 |
|---|
| BRCA1 |   | 5 | 148 | 598 | 575 | 0.157 | -0.201 | -0.078 | -0.087 | 0.168 |
|---|
| TCF7L2 |   | 17 | 30 | 271 | 264 | -0.071 | -0.021 | 0.143 | 0.280 | 0.101 |
|---|
| COX17 |   | 1 | 652 | 1472 | 1472 | -0.042 | 0.249 | -0.006 | -0.094 | 0.125 |
|---|
| PTGS2 |   | 2 | 391 | 1198 | 1184 | 0.022 | 0.091 | -0.073 | 0.250 | 0.152 |
|---|
| KPNA1 |   | 3 | 265 | 1198 | 1173 | 0.130 | 0.007 | 0.152 | -0.083 | 0.086 |
|---|
| CASP7 |   | 8 | 84 | 283 | 278 | -0.270 | 0.216 | 0.095 | 0.255 | 0.035 |
|---|
| IKBKE |   | 8 | 84 | 756 | 726 | -0.033 | 0.227 | -0.041 | -0.095 | 0.079 |
|---|
| RAD51 |   | 4 | 198 | 696 | 684 | -0.225 | 0.146 | -0.098 | 0.135 | 0.061 |
|---|
| PITX1 |   | 2 | 391 | 1410 | 1391 | -0.105 | 0.256 | 0.046 | 0.091 | 0.139 |
|---|
| GRIN2D |   | 18 | 26 | 33 | 32 | undef | 0.058 | 0.155 | 0.000 | undef |
|---|
| ESR1 |   | 4 | 198 | 1292 | 1277 | 0.046 | 0.026 | -0.002 | -0.053 | -0.062 |
|---|
| FOSL1 |   | 2 | 391 | 606 | 609 | 0.034 | -0.040 | 0.164 | 0.010 | 0.005 |
|---|
| GNA13 |   | 3 | 265 | 910 | 895 | -0.103 | 0.109 | -0.208 | 0.108 | -0.092 |
|---|
| COPG |   | 2 | 391 | 696 | 698 | -0.040 | 0.004 | 0.080 | -0.158 | 0.280 |
|---|
GO Enrichment output for subnetwork 427 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein sumoylation | GO:0016925 |  | 1.239E-12 | 2.851E-09 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 2.413E-07 | 2.775E-04 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 8.164E-07 | 6.259E-04 |
|---|
| protein modification by small protein conjugation | GO:0032446 |  | 2.781E-06 | 1.599E-03 |
|---|
| protein modification by small protein conjugation or removal | GO:0070647 |  | 4.055E-06 | 1.865E-03 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 4.528E-06 | 1.736E-03 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 5.549E-06 | 1.823E-03 |
|---|
| negative regulation of transcription regulator activity | GO:0090048 |  | 5.653E-06 | 1.625E-03 |
|---|
| intracellular receptor-mediated signaling pathway | GO:0030522 |  | 8.368E-06 | 2.138E-03 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 9.046E-06 | 2.08E-03 |
|---|
| negative regulation of DNA binding | GO:0043392 |  | 1.292E-05 | 2.701E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein sumoylation | GO:0016925 |  | 1.681E-13 | 4.108E-10 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.234E-10 | 1.508E-07 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 1.345E-09 | 1.095E-06 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 6.312E-08 | 3.855E-05 |
|---|
| intracellular receptor-mediated signaling pathway | GO:0030522 |  | 9.825E-08 | 4.8E-05 |
|---|
| myoblast differentiation | GO:0045445 |  | 2.078E-07 | 8.459E-05 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 2.665E-07 | 9.3E-05 |
|---|
| Notch receptor processing | GO:0007220 |  | 2.951E-07 | 9.01E-05 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 2.951E-07 | 8.009E-05 |
|---|
| skeletal muscle fiber development | GO:0048741 |  | 9.125E-07 | 2.229E-04 |
|---|
| muscle fiber development | GO:0048747 |  | 9.977E-07 | 2.216E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein sumoylation | GO:0016925 |  | 2.541E-13 | 6.115E-10 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 9.543E-11 | 1.148E-07 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 1.778E-09 | 1.426E-06 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 7.124E-08 | 4.285E-05 |
|---|
| intracellular receptor-mediated signaling pathway | GO:0030522 |  | 1.132E-07 | 5.448E-05 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 2.742E-07 | 1.099E-04 |
|---|
| myoblast differentiation | GO:0045445 |  | 2.742E-07 | 9.424E-05 |
|---|
| Notch receptor processing | GO:0007220 |  | 3.643E-07 | 1.096E-04 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 3.643E-07 | 9.739E-05 |
|---|
| amyloid precursor protein catabolic process | GO:0042987 |  | 1.269E-06 | 3.053E-04 |
|---|
| skeletal muscle fiber development | GO:0048741 |  | 1.28E-06 | 2.8E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein sumoylation | GO:0016925 |  | 1.239E-12 | 2.851E-09 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 2.413E-07 | 2.775E-04 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 8.164E-07 | 6.259E-04 |
|---|
| protein modification by small protein conjugation | GO:0032446 |  | 2.781E-06 | 1.599E-03 |
|---|
| protein modification by small protein conjugation or removal | GO:0070647 |  | 4.055E-06 | 1.865E-03 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 4.528E-06 | 1.736E-03 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 5.549E-06 | 1.823E-03 |
|---|
| negative regulation of transcription regulator activity | GO:0090048 |  | 5.653E-06 | 1.625E-03 |
|---|
| intracellular receptor-mediated signaling pathway | GO:0030522 |  | 8.368E-06 | 2.138E-03 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 9.046E-06 | 2.08E-03 |
|---|
| negative regulation of DNA binding | GO:0043392 |  | 1.292E-05 | 2.701E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein sumoylation | GO:0016925 |  | 1.239E-12 | 2.851E-09 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 2.413E-07 | 2.775E-04 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 8.164E-07 | 6.259E-04 |
|---|
| protein modification by small protein conjugation | GO:0032446 |  | 2.781E-06 | 1.599E-03 |
|---|
| protein modification by small protein conjugation or removal | GO:0070647 |  | 4.055E-06 | 1.865E-03 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 4.528E-06 | 1.736E-03 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 5.549E-06 | 1.823E-03 |
|---|
| negative regulation of transcription regulator activity | GO:0090048 |  | 5.653E-06 | 1.625E-03 |
|---|
| intracellular receptor-mediated signaling pathway | GO:0030522 |  | 8.368E-06 | 2.138E-03 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 9.046E-06 | 2.08E-03 |
|---|
| negative regulation of DNA binding | GO:0043392 |  | 1.292E-05 | 2.701E-03 |
|---|


