Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1324
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3416 | 1.030e-03 | 1.400e-05 | 1.375e-01 | 1.983e-09 |
|---|
| IPC | 0.3040 | 4.086e-01 | 2.341e-01 | 9.478e-01 | 9.067e-02 |
|---|
| Loi | 0.4918 | 2.600e-05 | 0.000e+00 | 4.602e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5915 | 1.000e-06 | 0.000e+00 | 1.312e-01 | 0.000e+00 |
|---|
| Wang | 0.2520 | 9.456e-03 | 7.573e-02 | 2.534e-01 | 1.815e-04 |
|---|
Expression data for subnetwork 1324 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
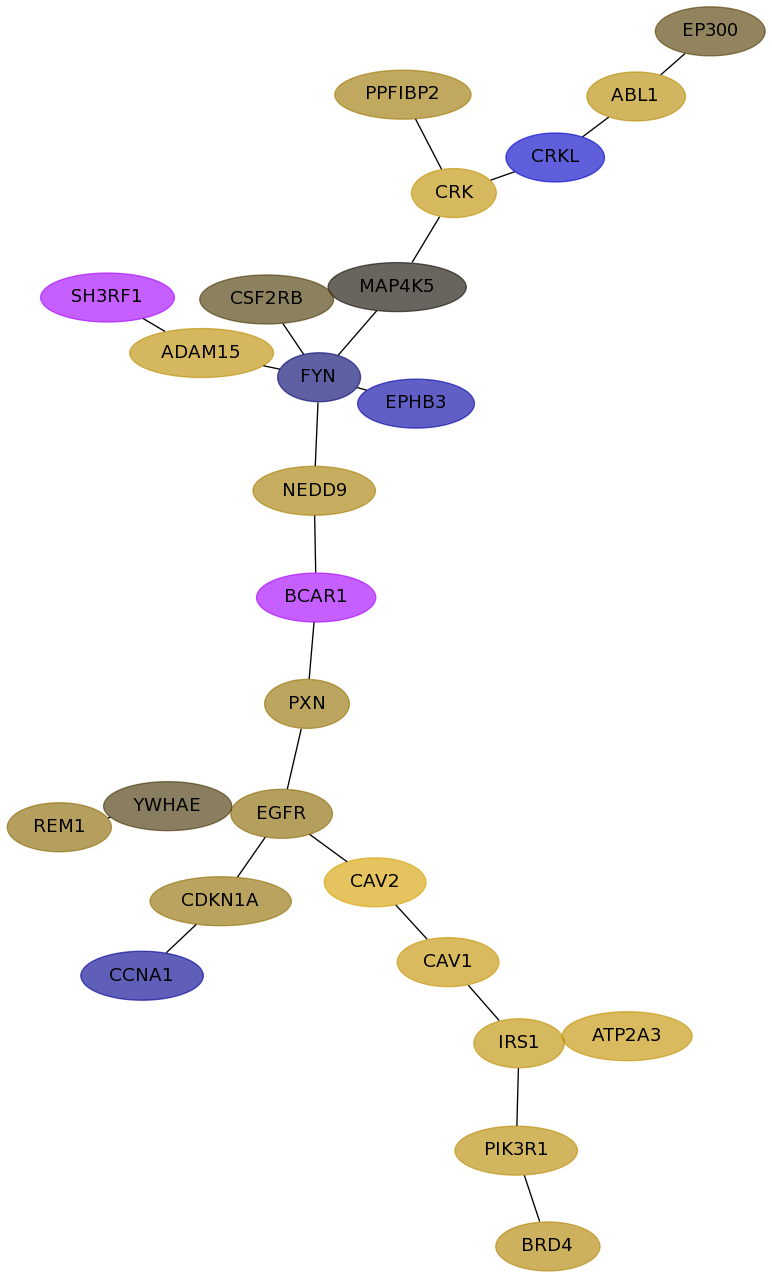
Score for each gene in subnetwork 1324 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| PXN |   | 5 | 148 | 439 | 432 | 0.085 | -0.088 | 0.284 | -0.022 | 0.114 |
|---|
| BRD4 |   | 18 | 26 | 1 | 12 | 0.156 | -0.119 | 0.095 | -0.075 | -0.044 |
|---|
| NEDD9 |   | 2 | 391 | 1052 | 1044 | 0.126 | 0.020 | 0.023 | 0.032 | 0.222 |
|---|
| ATP2A3 |   | 9 | 77 | 1 | 18 | 0.240 | -0.040 | 0.082 | -0.229 | -0.025 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| PPFIBP2 |   | 7 | 100 | 606 | 576 | 0.098 | 0.188 | 0.320 | -0.112 | 0.013 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| CAV1 |   | 10 | 67 | 439 | 428 | 0.251 | -0.092 | 0.124 | 0.079 | 0.119 |
|---|
| YWHAE |   | 1 | 652 | 1391 | 1392 | 0.012 | -0.018 | 0.112 | 0.279 | 0.014 |
|---|
| BCAR1 |   | 1 | 652 | 1391 | 1392 | undef | -0.012 | 0.229 | undef | undef |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| IRS1 |   | 14 | 42 | 1 | 14 | 0.217 | -0.219 | 0.065 | 0.165 | 0.064 |
|---|
| CRKL |   | 7 | 100 | 326 | 317 | -0.271 | -0.023 | 0.056 | 0.271 | -0.008 |
|---|
| ABL1 |   | 8 | 84 | 326 | 316 | 0.193 | -0.152 | 0.200 | 0.108 | -0.059 |
|---|
| CSF2RB |   | 3 | 265 | 691 | 685 | 0.013 | 0.013 | -0.196 | -0.109 | -0.141 |
|---|
| FYN |   | 3 | 265 | 182 | 189 | -0.032 | -0.171 | -0.167 | 0.073 | 0.110 |
|---|
| CRK |   | 27 | 17 | 206 | 193 | 0.232 | -0.116 | 0.106 | 0.090 | 0.017 |
|---|
| MAP4K5 |   | 2 | 391 | 412 | 423 | 0.002 | 0.103 | 0.202 | 0.028 | 0.061 |
|---|
| SH3RF1 |   | 14 | 42 | 117 | 120 | undef | 0.144 | 0.238 | undef | undef |
|---|
| CCNA1 |   | 17 | 30 | 326 | 314 | -0.078 | 0.174 | 0.092 | 0.078 | -0.098 |
|---|
| ADAM15 |   | 13 | 47 | 492 | 474 | 0.205 | -0.048 | -0.078 | -0.208 | -0.048 |
|---|
| REM1 |   | 1 | 652 | 1391 | 1392 | 0.064 | 0.106 | 0.136 | -0.067 | -0.122 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| EPHB3 |   | 4 | 198 | 326 | 333 | -0.115 | -0.048 | 0.135 | 0.099 | -0.008 |
|---|
GO Enrichment output for subnetwork 1324 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 1.285E-07 | 2.956E-04 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 1.285E-07 | 1.478E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.356E-07 | 1.039E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 2.566E-07 | 1.475E-04 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 2.566E-07 | 1.18E-04 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 2.566E-07 | 9.836E-05 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 4.483E-07 | 1.473E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 7.161E-07 | 2.059E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 7.161E-07 | 1.83E-04 |
|---|
| mammary gland development | GO:0030879 |  | 8.637E-07 | 1.987E-04 |
|---|
| regulation of epithelial cell differentiation | GO:0030856 |  | 1.072E-06 | 2.242E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 6.384E-08 | 1.56E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 6.735E-08 | 8.226E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.958E-07 | 1.595E-04 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 2.228E-07 | 1.361E-04 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 2.228E-07 | 1.089E-04 |
|---|
| regulation of MAPKKK cascade | GO:0043408 |  | 2.594E-07 | 1.056E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 3.561E-07 | 1.243E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 3.561E-07 | 1.087E-04 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 3.561E-07 | 9.665E-05 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 3.561E-07 | 8.699E-05 |
|---|
| regulation of epithelial cell differentiation | GO:0030856 |  | 5.334E-07 | 1.185E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 6.731E-08 | 1.62E-04 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 7.496E-08 | 9.017E-05 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 1.497E-07 | 1.201E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.059E-07 | 1.238E-04 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 2.616E-07 | 1.259E-04 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 2.616E-07 | 1.049E-04 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 2.616E-07 | 8.992E-05 |
|---|
| regulation of MAPKKK cascade | GO:0043408 |  | 3.116E-07 | 9.37E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 4.18E-07 | 1.117E-04 |
|---|
| regulation of epithelial cell differentiation | GO:0030856 |  | 6.261E-07 | 1.506E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 6.261E-07 | 1.369E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 1.285E-07 | 2.956E-04 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 1.285E-07 | 1.478E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.356E-07 | 1.039E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 2.566E-07 | 1.475E-04 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 2.566E-07 | 1.18E-04 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 2.566E-07 | 9.836E-05 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 4.483E-07 | 1.473E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 7.161E-07 | 2.059E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 7.161E-07 | 1.83E-04 |
|---|
| mammary gland development | GO:0030879 |  | 8.637E-07 | 1.987E-04 |
|---|
| regulation of epithelial cell differentiation | GO:0030856 |  | 1.072E-06 | 2.242E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 1.285E-07 | 2.956E-04 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 1.285E-07 | 1.478E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.356E-07 | 1.039E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 2.566E-07 | 1.475E-04 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 2.566E-07 | 1.18E-04 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 2.566E-07 | 9.836E-05 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 4.483E-07 | 1.473E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 7.161E-07 | 2.059E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 7.161E-07 | 1.83E-04 |
|---|
| mammary gland development | GO:0030879 |  | 8.637E-07 | 1.987E-04 |
|---|
| regulation of epithelial cell differentiation | GO:0030856 |  | 1.072E-06 | 2.242E-04 |
|---|


