Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1308
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3399 | 1.117e-03 | 1.600e-05 | 1.425e-01 | 2.547e-09 |
|---|
| IPC | 0.3075 | 4.031e-01 | 2.287e-01 | 9.467e-01 | 8.728e-02 |
|---|
| Loi | 0.4903 | 2.800e-05 | 0.000e+00 | 4.671e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5930 | 1.000e-06 | 0.000e+00 | 1.278e-01 | 0.000e+00 |
|---|
| Wang | 0.2525 | 9.270e-03 | 7.479e-02 | 2.512e-01 | 1.741e-04 |
|---|
Expression data for subnetwork 1308 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
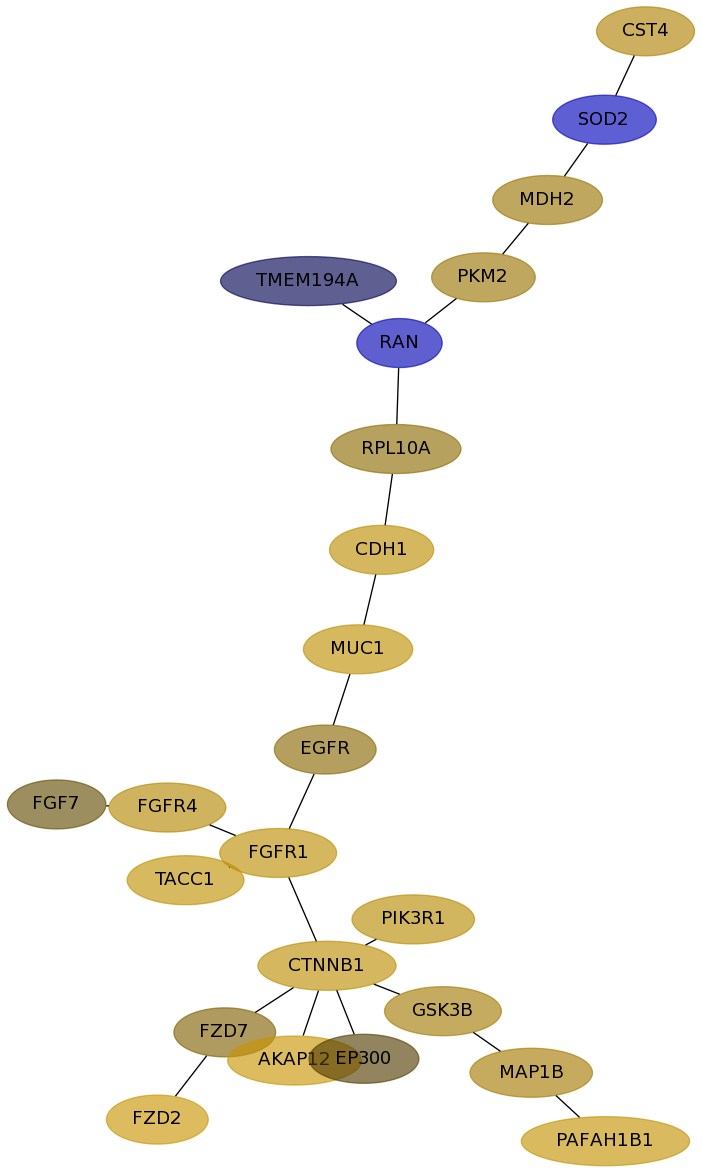
Score for each gene in subnetwork 1308 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| TACC1 |   | 5 | 148 | 326 | 330 | 0.229 | -0.051 | 0.020 | -0.022 | 0.126 |
|---|
| PAFAH1B1 |   | 1 | 652 | 1329 | 1330 | 0.239 | -0.118 | 0.130 | 0.146 | 0.074 |
|---|
| CST4 |   | 1 | 652 | 1329 | 1330 | 0.152 | 0.070 | 0.336 | -0.033 | 0.068 |
|---|
| MDH2 |   | 1 | 652 | 1329 | 1330 | 0.093 | 0.089 | 0.180 | 0.044 | -0.008 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| SOD2 |   | 1 | 652 | 1329 | 1330 | -0.206 | 0.176 | -0.056 | -0.051 | -0.019 |
|---|
| AKAP12 |   | 1 | 652 | 1329 | 1330 | 0.275 | -0.107 | 0.095 | 0.104 | 0.276 |
|---|
| PKM2 |   | 1 | 652 | 1329 | 1330 | 0.093 | -0.018 | 0.257 | 0.056 | -0.050 |
|---|
| RPL10A |   | 2 | 391 | 1135 | 1135 | 0.068 | 0.176 | 0.061 | 0.219 | -0.024 |
|---|
| FZD2 |   | 8 | 84 | 552 | 540 | 0.273 | -0.067 | -0.081 | -0.067 | 0.081 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| FZD7 |   | 5 | 148 | 552 | 541 | 0.051 | -0.219 | 0.062 | 0.062 | -0.059 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| MAP1B |   | 13 | 47 | 148 | 148 | 0.118 | -0.123 | 0.263 | -0.107 | 0.100 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| TMEM194A |   | 1 | 652 | 1329 | 1330 | -0.017 | -0.015 | -0.161 | -0.007 | 0.087 |
|---|
| FGF7 |   | 8 | 84 | 398 | 384 | 0.026 | -0.094 | 0.237 | 0.070 | 0.083 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| RAN |   | 2 | 391 | 715 | 711 | -0.164 | 0.030 | -0.006 | 0.062 | undef |
|---|
GO Enrichment output for subnetwork 1308 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 6.031E-07 | 1.387E-03 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.22E-06 | 1.403E-03 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 4.827E-06 | 3.701E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.455E-05 | 8.368E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 2.065E-05 | 9.501E-03 |
|---|
| malate metabolic process | GO:0006108 |  | 7.973E-05 | 0.03056317 |
|---|
| cerebral cortex radially oriented cell migration | GO:0021799 |  | 7.973E-05 | 0.026197 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 9.652E-05 | 0.02775005 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.131E-04 | 0.02889353 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.194E-04 | 0.02745653 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 1.194E-04 | 0.02496048 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 5.091E-09 | 1.244E-05 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 6.997E-09 | 8.547E-06 |
|---|
| cerebral cortex radially oriented cell migration | GO:0021799 |  | 2.023E-07 | 1.648E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 2.023E-07 | 1.236E-04 |
|---|
| platelet activating factor metabolic process | GO:0046469 |  | 3.536E-07 | 1.727E-04 |
|---|
| cerebral cortex cell migration | GO:0021795 |  | 8.458E-07 | 3.444E-04 |
|---|
| brain morphogenesis | GO:0048854 |  | 1.206E-06 | 4.21E-04 |
|---|
| central nervous system neuron axonogenesis | GO:0021955 |  | 1.206E-06 | 3.684E-04 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.341E-06 | 3.64E-04 |
|---|
| telencephalon cell migration | GO:0022029 |  | 1.656E-06 | 4.046E-04 |
|---|
| neuroblast proliferation | GO:0007405 |  | 1.656E-06 | 3.678E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 4.939E-09 | 1.188E-05 |
|---|
| cerebral cortex radially oriented cell migration | GO:0021799 |  | 2.191E-07 | 2.636E-04 |
|---|
| establishment of spindle localization | GO:0051293 |  | 2.191E-07 | 1.757E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.9E-07 | 1.745E-04 |
|---|
| platelet activating factor metabolic process | GO:0046469 |  | 3.828E-07 | 1.842E-04 |
|---|
| cerebral cortex cell migration | GO:0021795 |  | 9.158E-07 | 3.672E-04 |
|---|
| central nervous system neuron axonogenesis | GO:0021955 |  | 1.306E-06 | 4.489E-04 |
|---|
| brain morphogenesis | GO:0048854 |  | 1.306E-06 | 3.928E-04 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.326E-06 | 3.544E-04 |
|---|
| telencephalon cell migration | GO:0022029 |  | 1.793E-06 | 4.314E-04 |
|---|
| neuroblast proliferation | GO:0007405 |  | 1.793E-06 | 3.922E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 6.031E-07 | 1.387E-03 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.22E-06 | 1.403E-03 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 4.827E-06 | 3.701E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.455E-05 | 8.368E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 2.065E-05 | 9.501E-03 |
|---|
| malate metabolic process | GO:0006108 |  | 7.973E-05 | 0.03056317 |
|---|
| cerebral cortex radially oriented cell migration | GO:0021799 |  | 7.973E-05 | 0.026197 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 9.652E-05 | 0.02775005 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.131E-04 | 0.02889353 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.194E-04 | 0.02745653 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 1.194E-04 | 0.02496048 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 6.031E-07 | 1.387E-03 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.22E-06 | 1.403E-03 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 4.827E-06 | 3.701E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.455E-05 | 8.368E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 2.065E-05 | 9.501E-03 |
|---|
| malate metabolic process | GO:0006108 |  | 7.973E-05 | 0.03056317 |
|---|
| cerebral cortex radially oriented cell migration | GO:0021799 |  | 7.973E-05 | 0.026197 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 9.652E-05 | 0.02775005 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.131E-04 | 0.02889353 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.194E-04 | 0.02745653 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 1.194E-04 | 0.02496048 |
|---|


