Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1302
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3388 | 1.177e-03 | 1.800e-05 | 1.458e-01 | 3.089e-09 |
|---|
| IPC | 0.3104 | 3.985e-01 | 2.242e-01 | 9.457e-01 | 8.448e-02 |
|---|
| Loi | 0.4893 | 3.000e-05 | 0.000e+00 | 4.717e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5946 | 1.000e-06 | 0.000e+00 | 1.243e-01 | 0.000e+00 |
|---|
| Wang | 0.2530 | 9.082e-03 | 7.384e-02 | 2.488e-01 | 1.669e-04 |
|---|
Expression data for subnetwork 1302 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
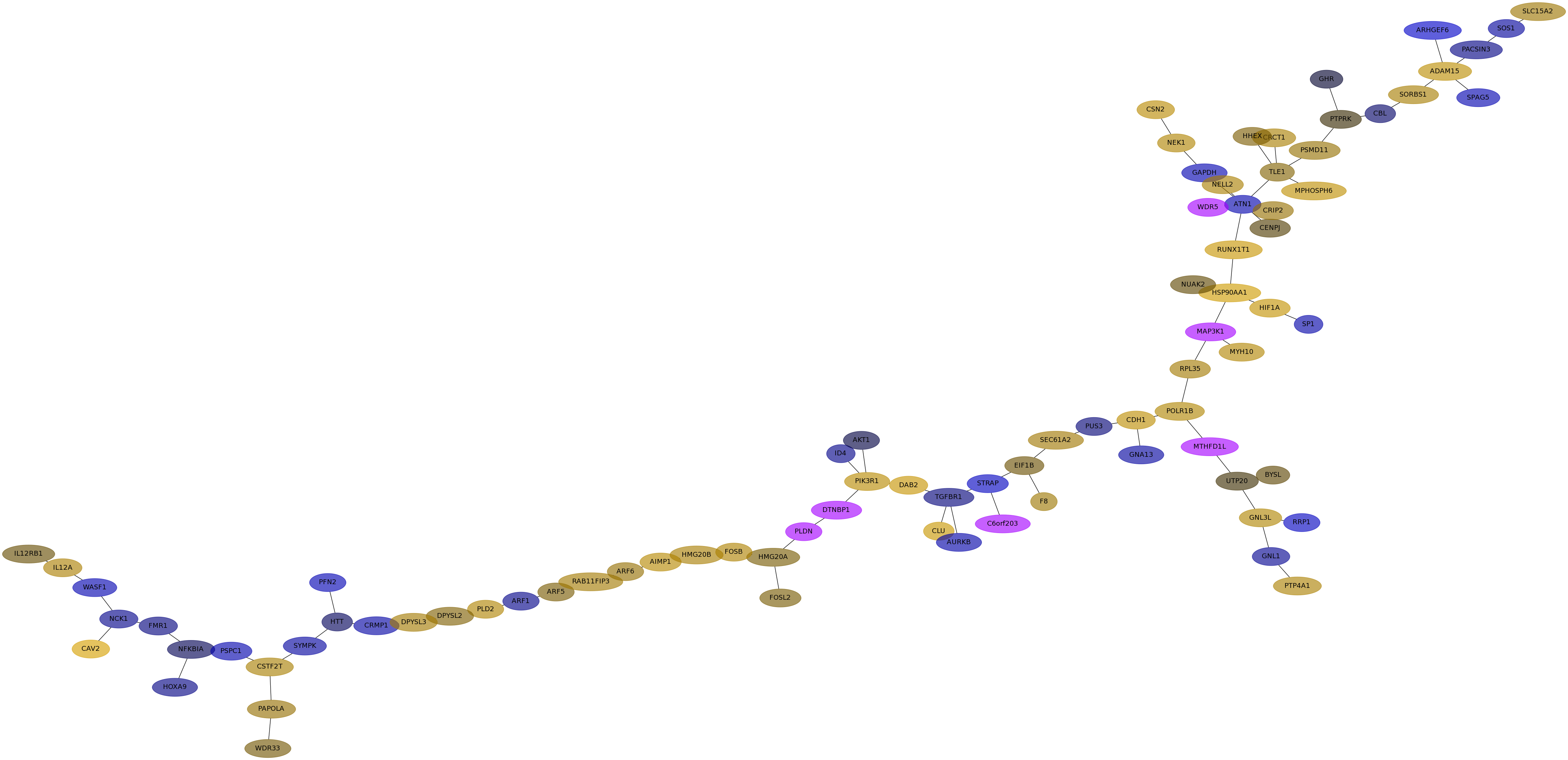
Score for each gene in subnetwork 1302 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| PTP4A1 |   | 1 | 652 | 1236 | 1238 | 0.142 | 0.216 | 0.023 | 0.034 | 0.239 |
|---|
| SORBS1 |   | 17 | 30 | 182 | 178 | 0.124 | -0.043 | 0.015 | 0.314 | 0.200 |
|---|
| WASF1 |   | 2 | 391 | 182 | 195 | -0.159 | 0.162 | -0.070 | 0.348 | 0.130 |
|---|
| ID4 |   | 13 | 47 | 444 | 430 | -0.060 | -0.179 | 0.137 | 0.241 | 0.036 |
|---|
| FOSL2 |   | 1 | 652 | 1236 | 1238 | 0.041 | 0.100 | 0.080 | -0.119 | 0.080 |
|---|
| MYH10 |   | 2 | 391 | 1236 | 1221 | 0.163 | -0.029 | 0.139 | 0.132 | 0.223 |
|---|
| EIF1B |   | 6 | 120 | 57 | 62 | 0.029 | 0.149 | 0.075 | 0.332 | -0.070 |
|---|
| PAPOLA |   | 1 | 652 | 1236 | 1238 | 0.081 | -0.042 | -0.072 | 0.179 | 0.168 |
|---|
| RAB11FIP3 |   | 1 | 652 | 1236 | 1238 | 0.116 | -0.038 | -0.016 | 0.047 | -0.043 |
|---|
| NELL2 |   | 2 | 391 | 444 | 458 | 0.134 | -0.005 | 0.138 | 0.421 | 0.074 |
|---|
| IL12A |   | 1 | 652 | 1236 | 1238 | 0.136 | 0.140 | -0.101 | 0.096 | 0.131 |
|---|
| PUS3 |   | 1 | 652 | 1236 | 1238 | -0.039 | 0.022 | 0.054 | 0.085 | 0.134 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CRCT1 |   | 1 | 652 | 1236 | 1238 | 0.133 | 0.135 | 0.050 | 0.000 | 0.154 |
|---|
| DPYSL2 |   | 3 | 265 | 148 | 166 | 0.050 | -0.095 | -0.037 | 0.296 | 0.104 |
|---|
| FOSB |   | 1 | 652 | 1236 | 1238 | 0.149 | -0.033 | 0.046 | 0.129 | 0.095 |
|---|
| DTNBP1 |   | 1 | 652 | 1236 | 1238 | undef | 0.210 | -0.071 | undef | undef |
|---|
| STRAP |   | 1 | 652 | 1236 | 1238 | -0.232 | 0.035 | -0.128 | 0.074 | -0.049 |
|---|
| SEC61A2 |   | 3 | 265 | 271 | 280 | 0.105 | -0.035 | -0.072 | -0.115 | -0.073 |
|---|
| PTPRK |   | 6 | 120 | 148 | 156 | 0.008 | 0.156 | 0.141 | -0.094 | -0.024 |
|---|
| RPL35 |   | 5 | 148 | 57 | 63 | 0.114 | 0.108 | -0.012 | 0.370 | -0.220 |
|---|
| GAPDH |   | 2 | 391 | 563 | 567 | -0.153 | 0.159 | -0.040 | -0.053 | 0.029 |
|---|
| CSN2 |   | 1 | 652 | 1236 | 1238 | 0.195 | -0.036 | 0.177 | -0.121 | -0.011 |
|---|
| AKT1 |   | 13 | 47 | 444 | 430 | -0.012 | 0.045 | 0.126 | -0.099 | -0.168 |
|---|
| RRP1 |   | 2 | 391 | 1008 | 999 | -0.224 | 0.154 | -0.104 | 0.095 | -0.135 |
|---|
| RUNX1T1 |   | 3 | 265 | 536 | 537 | 0.271 | -0.170 | -0.040 | 0.081 | -0.099 |
|---|
| NCK1 |   | 3 | 265 | 206 | 208 | -0.065 | 0.135 | -0.100 | -0.059 | 0.151 |
|---|
| GNL1 |   | 1 | 652 | 1236 | 1238 | -0.074 | 0.076 | 0.078 | 0.303 | 0.086 |
|---|
| AURKB |   | 3 | 265 | 1236 | 1219 | -0.132 | 0.179 | -0.189 | 0.086 | -0.028 |
|---|
| MTHFD1L |   | 1 | 652 | 1236 | 1238 | undef | -0.004 | -0.042 | undef | undef |
|---|
| IL12RB1 |   | 1 | 652 | 1236 | 1238 | 0.027 | 0.175 | -0.177 | -0.041 | -0.034 |
|---|
| CBL |   | 1 | 652 | 1236 | 1238 | -0.028 | 0.031 | 0.023 | 0.164 | -0.126 |
|---|
| DPYSL3 |   | 2 | 391 | 148 | 171 | 0.113 | -0.065 | 0.133 | 0.021 | 0.261 |
|---|
| HHEX |   | 1 | 652 | 1236 | 1238 | 0.045 | -0.130 | -0.178 | 0.177 | 0.005 |
|---|
| C6orf203 |   | 1 | 652 | 1236 | 1238 | undef | 0.285 | -0.074 | undef | undef |
|---|
| ARHGEF6 |   | 3 | 265 | 859 | 843 | -0.274 | -0.068 | -0.120 | 0.166 | -0.115 |
|---|
| HOXA9 |   | 2 | 391 | 634 | 628 | -0.055 | 0.057 | 0.136 | 0.040 | 0.018 |
|---|
| TLE1 |   | 2 | 391 | 1177 | 1166 | 0.054 | 0.183 | 0.050 | 0.030 | 0.055 |
|---|
| GNL3L |   | 3 | 265 | 994 | 971 | 0.158 | 0.140 | 0.020 | -0.264 | -0.031 |
|---|
| CLU |   | 10 | 67 | 117 | 121 | 0.280 | -0.208 | 0.119 | -0.153 | -0.234 |
|---|
| ARF6 |   | 1 | 652 | 1236 | 1238 | 0.081 | 0.211 | -0.044 | 0.066 | -0.242 |
|---|
| GNA13 |   | 3 | 265 | 910 | 895 | -0.103 | 0.109 | -0.208 | 0.108 | -0.092 |
|---|
| CRMP1 |   | 3 | 265 | 647 | 632 | -0.114 | -0.068 | 0.174 | 0.049 | 0.151 |
|---|
| F8 |   | 2 | 391 | 57 | 80 | 0.102 | 0.059 | -0.037 | 0.088 | 0.039 |
|---|
| CSTF2T |   | 1 | 652 | 1236 | 1238 | 0.130 | -0.102 | -0.245 | 0.140 | 0.109 |
|---|
| TGFBR1 |   | 1 | 652 | 1236 | 1238 | -0.045 | -0.004 | 0.036 | -0.078 | undef |
|---|
| PLD2 |   | 1 | 652 | 1236 | 1238 | 0.154 | -0.138 | 0.011 | 0.177 | -0.212 |
|---|
| ARF1 |   | 1 | 652 | 1236 | 1238 | -0.060 | 0.003 | 0.060 | -0.126 | 0.068 |
|---|
| WDR5 |   | 1 | 652 | 1236 | 1238 | undef | 0.099 | -0.000 | undef | undef |
|---|
| PFN2 |   | 4 | 198 | 182 | 183 | -0.164 | 0.014 | -0.001 | 0.286 | -0.056 |
|---|
| HMG20A |   | 1 | 652 | 1236 | 1238 | 0.041 | 0.043 | 0.055 | 0.154 | 0.001 |
|---|
| SPAG5 |   | 4 | 198 | 492 | 499 | -0.159 | 0.132 | -0.243 | 0.031 | 0.084 |
|---|
| DAB2 |   | 13 | 47 | 1 | 15 | 0.258 | -0.175 | 0.067 | -0.022 | 0.071 |
|---|
| CENPJ |   | 1 | 652 | 1236 | 1238 | 0.016 | 0.125 | -0.153 | 0.181 | 0.117 |
|---|
| ARF5 |   | 1 | 652 | 1236 | 1238 | 0.042 | 0.038 | -0.049 | -0.091 | 0.000 |
|---|
| GHR |   | 14 | 42 | 148 | 146 | -0.006 | -0.112 | 0.174 | 0.090 | 0.201 |
|---|
| ADAM15 |   | 13 | 47 | 492 | 474 | 0.205 | -0.048 | -0.078 | -0.208 | -0.048 |
|---|
| PSMD11 |   | 2 | 391 | 444 | 458 | 0.085 | 0.088 | -0.090 | -0.137 | -0.078 |
|---|
| HSP90AA1 |   | 10 | 67 | 1 | 16 | 0.321 | -0.143 | 0.104 | 0.123 | -0.054 |
|---|
| POLR1B |   | 11 | 59 | 129 | 126 | 0.157 | 0.062 | -0.036 | -0.079 | 0.139 |
|---|
| FMR1 |   | 3 | 265 | 679 | 667 | -0.047 | 0.137 | -0.104 | 0.352 | 0.113 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| MPHOSPH6 |   | 8 | 84 | 756 | 726 | 0.217 | -0.084 | 0.175 | -0.269 | 0.146 |
|---|
| MAP3K1 |   | 1 | 652 | 1236 | 1238 | undef | 0.112 | 0.039 | undef | undef |
|---|
| SP1 |   | 16 | 34 | 283 | 272 | -0.129 | 0.222 | -0.254 | 0.239 | 0.082 |
|---|
| SOS1 |   | 1 | 652 | 1236 | 1238 | -0.096 | -0.049 | 0.002 | -0.007 | -0.042 |
|---|
| PSPC1 |   | 2 | 391 | 679 | 678 | -0.148 | 0.203 | -0.289 | 0.298 | -0.107 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| CRIP2 |   | 1 | 652 | 1236 | 1238 | 0.085 | 0.104 | 0.069 | -0.081 | -0.098 |
|---|
| PACSIN3 |   | 3 | 265 | 778 | 770 | -0.055 | 0.270 | 0.129 | -0.045 | 0.066 |
|---|
| NUAK2 |   | 1 | 652 | 1236 | 1238 | 0.023 | 0.107 | -0.053 | -0.137 | 0.087 |
|---|
| HMG20B |   | 1 | 652 | 1236 | 1238 | 0.128 | -0.045 | 0.146 | -0.151 | -0.155 |
|---|
| PLDN |   | 1 | 652 | 1236 | 1238 | undef | 0.083 | -0.030 | undef | undef |
|---|
| SYMPK |   | 1 | 652 | 1236 | 1238 | -0.103 | 0.007 | -0.116 | -0.173 | 0.074 |
|---|
| SLC15A2 |   | 1 | 652 | 1236 | 1238 | 0.100 | -0.076 | 0.109 | -0.068 | -0.123 |
|---|
| UTP20 |   | 1 | 652 | 1236 | 1238 | 0.010 | -0.082 | -0.162 | 0.039 | -0.148 |
|---|
| NEK1 |   | 4 | 198 | 57 | 68 | 0.155 | 0.032 | 0.184 | 0.049 | 0.135 |
|---|
| HIF1A |   | 21 | 23 | 33 | 31 | 0.246 | -0.098 | 0.224 | -0.118 | 0.356 |
|---|
| HTT |   | 2 | 391 | 532 | 543 | -0.020 | -0.016 | -0.015 | 0.075 | -0.093 |
|---|
| NFKBIA |   | 2 | 391 | 679 | 678 | -0.018 | 0.133 | -0.146 | 0.007 | -0.334 |
|---|
| ATN1 |   | 2 | 391 | 444 | 458 | -0.149 | -0.005 | -0.093 | 0.005 | -0.136 |
|---|
| AIMP1 |   | 1 | 652 | 1236 | 1238 | 0.177 | 0.211 | -0.021 | 0.016 | 0.236 |
|---|
| WDR33 |   | 1 | 652 | 1236 | 1238 | 0.037 | 0.174 | -0.117 | 0.001 | 0.083 |
|---|
| BYSL |   | 1 | 652 | 1236 | 1238 | 0.021 | 0.064 | -0.000 | 0.108 | -0.103 |
|---|
GO Enrichment output for subnetwork 1302 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.936E-07 | 4.453E-04 |
|---|
| positive regulation of protein polymerization | GO:0032273 |  | 3.465E-07 | 3.984E-04 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 1.922E-06 | 1.473E-03 |
|---|
| negative regulation of endocytosis | GO:0045806 |  | 7.725E-06 | 4.442E-03 |
|---|
| positive regulation of nitric-oxide synthase activity | GO:0051000 |  | 7.725E-06 | 3.553E-03 |
|---|
| gas homeostasis | GO:0033483 |  | 7.725E-06 | 2.961E-03 |
|---|
| positive regulation of cytoskeleton organization | GO:0051495 |  | 7.992E-06 | 2.626E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.251E-05 | 3.596E-03 |
|---|
| positive regulation of monooxygenase activity | GO:0032770 |  | 1.344E-05 | 3.436E-03 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 2.14E-05 | 4.921E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.682E-05 | 5.607E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of actin filament polymerization | GO:0030838 |  | 4.508E-11 | 1.101E-07 |
|---|
| positive regulation of protein polymerization | GO:0032273 |  | 1.281E-10 | 1.564E-07 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 7.76E-10 | 6.319E-07 |
|---|
| positive regulation of cytoskeleton organization | GO:0051495 |  | 5.459E-09 | 3.334E-06 |
|---|
| positive regulation of organelle organization | GO:0010638 |  | 6.023E-08 | 2.943E-05 |
|---|
| negative regulation of endocytosis | GO:0045806 |  | 1.572E-07 | 6.401E-05 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.572E-07 | 5.487E-05 |
|---|
| regulation of actin polymerization or depolymerization | GO:0008064 |  | 1.006E-06 | 3.073E-04 |
|---|
| regulation of protein polymerization | GO:0032271 |  | 1.006E-06 | 2.732E-04 |
|---|
| regulation of actin filament length | GO:0030832 |  | 1.384E-06 | 3.382E-04 |
|---|
| regulation of cellular component size | GO:0032535 |  | 1.871E-06 | 4.155E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of actin filament polymerization | GO:0030838 |  | 6.968E-11 | 1.677E-07 |
|---|
| positive regulation of protein polymerization | GO:0032273 |  | 1.236E-10 | 1.487E-07 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 8.72E-10 | 6.994E-07 |
|---|
| positive regulation of cytoskeleton organization | GO:0051495 |  | 5.115E-09 | 3.077E-06 |
|---|
| positive regulation of organelle organization | GO:0010638 |  | 8.429E-08 | 4.056E-05 |
|---|
| negative regulation of endocytosis | GO:0045806 |  | 1.243E-07 | 4.983E-05 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.225E-07 | 7.649E-05 |
|---|
| regulation of actin polymerization or depolymerization | GO:0008064 |  | 1.329E-06 | 3.996E-04 |
|---|
| regulation of protein polymerization | GO:0032271 |  | 1.329E-06 | 3.552E-04 |
|---|
| regulation of actin filament length | GO:0030832 |  | 1.849E-06 | 4.448E-04 |
|---|
| regulation of cellular component size | GO:0032535 |  | 2.524E-06 | 5.52E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.936E-07 | 4.453E-04 |
|---|
| positive regulation of protein polymerization | GO:0032273 |  | 3.465E-07 | 3.984E-04 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 1.922E-06 | 1.473E-03 |
|---|
| negative regulation of endocytosis | GO:0045806 |  | 7.725E-06 | 4.442E-03 |
|---|
| positive regulation of nitric-oxide synthase activity | GO:0051000 |  | 7.725E-06 | 3.553E-03 |
|---|
| gas homeostasis | GO:0033483 |  | 7.725E-06 | 2.961E-03 |
|---|
| positive regulation of cytoskeleton organization | GO:0051495 |  | 7.992E-06 | 2.626E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.251E-05 | 3.596E-03 |
|---|
| positive regulation of monooxygenase activity | GO:0032770 |  | 1.344E-05 | 3.436E-03 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 2.14E-05 | 4.921E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.682E-05 | 5.607E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.936E-07 | 4.453E-04 |
|---|
| positive regulation of protein polymerization | GO:0032273 |  | 3.465E-07 | 3.984E-04 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 1.922E-06 | 1.473E-03 |
|---|
| negative regulation of endocytosis | GO:0045806 |  | 7.725E-06 | 4.442E-03 |
|---|
| positive regulation of nitric-oxide synthase activity | GO:0051000 |  | 7.725E-06 | 3.553E-03 |
|---|
| gas homeostasis | GO:0033483 |  | 7.725E-06 | 2.961E-03 |
|---|
| positive regulation of cytoskeleton organization | GO:0051495 |  | 7.992E-06 | 2.626E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.251E-05 | 3.596E-03 |
|---|
| positive regulation of monooxygenase activity | GO:0032770 |  | 1.344E-05 | 3.436E-03 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 2.14E-05 | 4.921E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.682E-05 | 5.607E-03 |
|---|


