Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1300
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3387 | 1.178e-03 | 1.800e-05 | 1.458e-01 | 3.093e-09 |
|---|
| IPC | 0.3094 | 4.000e-01 | 2.257e-01 | 9.460e-01 | 8.542e-02 |
|---|
| Loi | 0.4899 | 2.900e-05 | 0.000e+00 | 4.688e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5940 | 1.000e-06 | 0.000e+00 | 1.254e-01 | 0.000e+00 |
|---|
| Wang | 0.2531 | 9.043e-03 | 7.365e-02 | 2.484e-01 | 1.654e-04 |
|---|
Expression data for subnetwork 1300 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
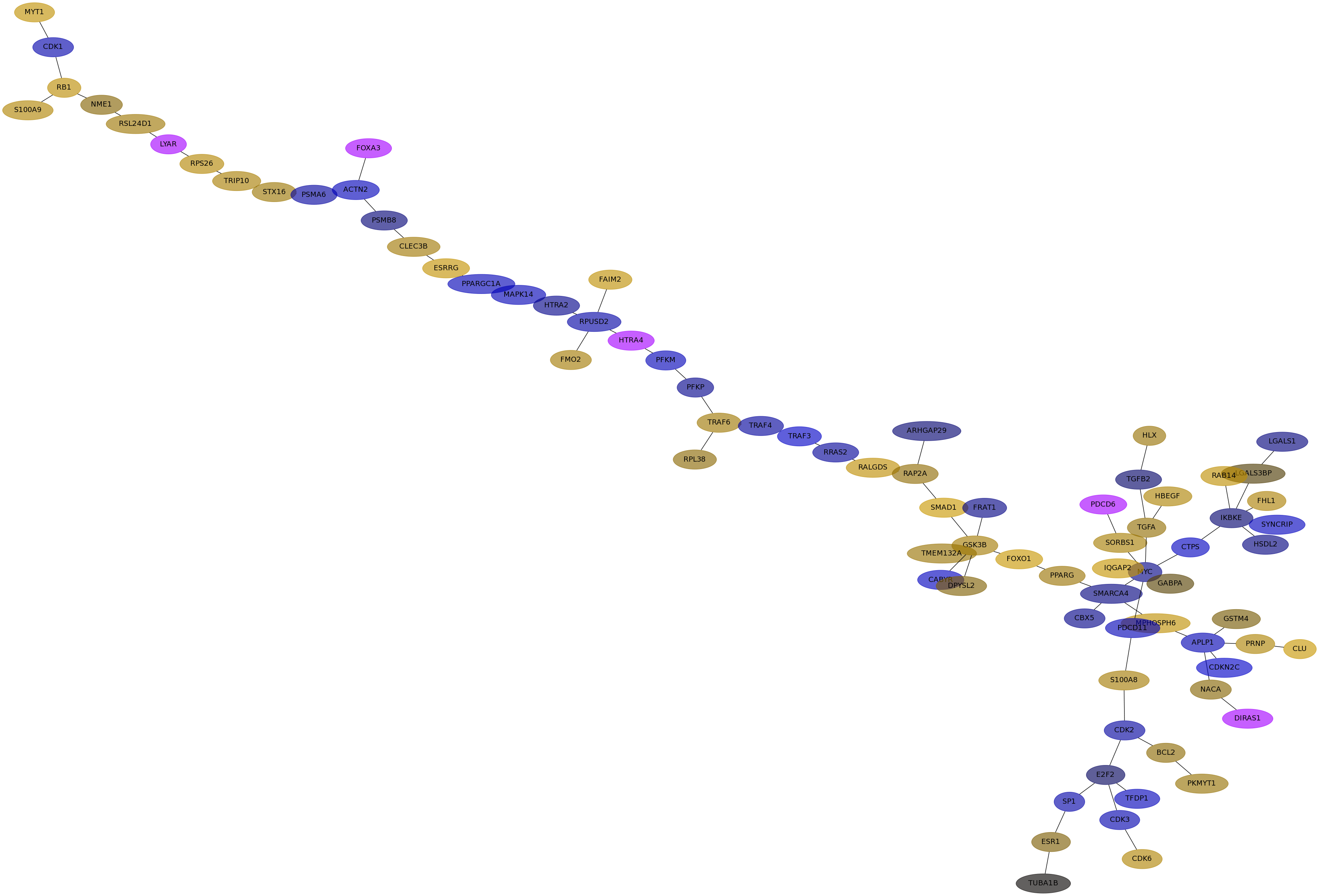
Score for each gene in subnetwork 1300 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| TRAF6 |   | 2 | 391 | 1106 | 1100 | 0.102 | -0.060 | -0.033 | 0.073 | -0.048 |
|---|
| RAB14 |   | 1 | 652 | 1292 | 1295 | 0.220 | -0.077 | 0.190 | 0.065 | -0.105 |
|---|
| SORBS1 |   | 17 | 30 | 182 | 178 | 0.124 | -0.043 | 0.015 | 0.314 | 0.200 |
|---|
| HBEGF |   | 1 | 652 | 1292 | 1295 | 0.149 | 0.087 | 0.002 | 0.039 | 0.076 |
|---|
| TRIP10 |   | 3 | 265 | 715 | 705 | 0.127 | 0.012 | 0.154 | 0.049 | 0.144 |
|---|
| SMARCA4 |   | 4 | 198 | 283 | 289 | -0.050 | 0.080 | 0.202 | -0.071 | -0.116 |
|---|
| S100A8 |   | 1 | 652 | 1292 | 1295 | 0.117 | 0.062 | -0.017 | -0.075 | 0.053 |
|---|
| TMEM132A |   | 7 | 100 | 33 | 37 | 0.088 | 0.019 | 0.127 | -0.141 | -0.092 |
|---|
| RPUSD2 |   | 2 | 391 | 1198 | 1184 | -0.123 | 0.209 | -0.015 | 0.199 | 0.169 |
|---|
| MAPK14 |   | 2 | 391 | 620 | 618 | -0.183 | 0.033 | -0.065 | 0.299 | 0.119 |
|---|
| CDK3 |   | 5 | 148 | 129 | 129 | -0.151 | -0.076 | 0.189 | -0.041 | 0.082 |
|---|
| PRNP |   | 1 | 652 | 1292 | 1295 | 0.147 | -0.090 | 0.168 | 0.020 | 0.048 |
|---|
| CDKN2C |   | 3 | 265 | 283 | 292 | -0.269 | 0.216 | -0.106 | 0.202 | -0.081 |
|---|
| PSMB8 |   | 4 | 198 | 710 | 696 | -0.039 | 0.254 | -0.071 | -0.088 | -0.139 |
|---|
| PSMA6 |   | 2 | 391 | 977 | 968 | -0.104 | 0.280 | -0.118 | -0.018 | -0.091 |
|---|
| HLX |   | 2 | 391 | 364 | 375 | 0.086 | 0.133 | -0.072 | 0.180 | 0.169 |
|---|
| DPYSL2 |   | 3 | 265 | 148 | 166 | 0.050 | -0.095 | -0.037 | 0.296 | 0.104 |
|---|
| RAP2A |   | 1 | 652 | 1292 | 1295 | 0.070 | -0.110 | 0.052 | 0.082 | -0.108 |
|---|
| MYT1 |   | 1 | 652 | 1292 | 1295 | 0.224 | -0.070 | -0.100 | 0.206 | -0.039 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
| HTRA2 |   | 2 | 391 | 859 | 860 | -0.063 | -0.123 | -0.118 | 0.234 | 0.011 |
|---|
| GABPA |   | 6 | 120 | 206 | 200 | 0.019 | 0.033 | -0.183 | 0.066 | -0.161 |
|---|
| PPARG |   | 3 | 265 | 283 | 292 | 0.092 | -0.119 | 0.011 | 0.105 | 0.039 |
|---|
| TGFA |   | 8 | 84 | 439 | 429 | 0.077 | -0.057 | -0.027 | -0.158 | 0.020 |
|---|
| LGALS1 |   | 3 | 265 | 696 | 686 | -0.045 | 0.267 | 0.010 | 0.218 | 0.156 |
|---|
| RPL38 |   | 3 | 265 | 859 | 843 | 0.065 | 0.222 | 0.151 | 0.241 | -0.171 |
|---|
| PKMYT1 |   | 5 | 148 | 620 | 603 | 0.085 | 0.175 | -0.143 | 0.046 | -0.070 |
|---|
| STX16 |   | 1 | 652 | 1292 | 1295 | 0.098 | -0.041 | 0.027 | 0.279 | 0.088 |
|---|
| CDK1 |   | 16 | 34 | 206 | 194 | -0.138 | 0.265 | -0.195 | 0.106 | 0.031 |
|---|
| TUBA1B |   | 7 | 100 | 182 | 180 | 0.001 | 0.116 | 0.094 | 0.249 | 0.128 |
|---|
| HSDL2 |   | 1 | 652 | 1292 | 1295 | -0.048 | -0.035 | -0.045 | 0.128 | 0.056 |
|---|
| NME1 |   | 7 | 100 | 492 | 495 | 0.054 | 0.267 | 0.106 | 0.046 | -0.035 |
|---|
| HTRA4 |   | 3 | 265 | 492 | 503 | undef | 0.102 | undef | undef | undef |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| TRAF3 |   | 1 | 652 | 1292 | 1295 | -0.260 | 0.019 | -0.208 | 0.234 | -0.025 |
|---|
| DIRAS1 |   | 1 | 652 | 1292 | 1295 | undef | 0.018 | 0.006 | undef | undef |
|---|
| CLU |   | 10 | 67 | 117 | 121 | 0.280 | -0.208 | 0.119 | -0.153 | -0.234 |
|---|
| ARHGAP29 |   | 1 | 652 | 1292 | 1295 | -0.034 | 0.187 | 0.096 | 0.258 | 0.155 |
|---|
| SMAD1 |   | 24 | 21 | 33 | 30 | 0.287 | 0.049 | 0.159 | -0.062 | 0.048 |
|---|
| PPARGC1A |   | 2 | 391 | 444 | 458 | -0.177 | 0.066 | -0.123 | 0.243 | 0.112 |
|---|
| TRAF4 |   | 1 | 652 | 1292 | 1295 | -0.097 | -0.181 | -0.091 | -0.071 | 0.196 |
|---|
| E2F2 |   | 4 | 198 | 129 | 130 | -0.021 | 0.212 | -0.047 | 0.162 | -0.092 |
|---|
| PDCD11 |   | 1 | 652 | 1292 | 1295 | -0.182 | -0.019 | 0.075 | 0.091 | 0.114 |
|---|
| RPS26 |   | 1 | 652 | 1292 | 1295 | 0.165 | 0.107 | -0.079 | 0.160 | -0.003 |
|---|
| PFKP |   | 1 | 652 | 1292 | 1295 | -0.066 | 0.195 | -0.076 | -0.040 | 0.094 |
|---|
| FAIM2 |   | 2 | 391 | 859 | 860 | 0.220 | 0.056 | 0.001 | -0.159 | -0.203 |
|---|
| FOXO1 |   | 4 | 198 | 148 | 162 | 0.278 | 0.072 | 0.094 | -0.021 | 0.221 |
|---|
| NACA |   | 1 | 652 | 1292 | 1295 | 0.058 | 0.075 | 0.158 | 0.046 | 0.252 |
|---|
| FOXA3 |   | 6 | 120 | 657 | 631 | undef | 0.192 | 0.289 | undef | undef |
|---|
| RALGDS |   | 5 | 148 | 647 | 621 | 0.212 | -0.011 | 0.199 | 0.033 | -0.009 |
|---|
| ACTN2 |   | 2 | 391 | 977 | 968 | -0.198 | 0.139 | -0.106 | 0.155 | 0.127 |
|---|
| TGFB2 |   | 7 | 100 | 57 | 60 | -0.028 | -0.063 | 0.244 | 0.226 | 0.015 |
|---|
| LGALS3BP |   | 1 | 652 | 1292 | 1295 | 0.015 | 0.167 | 0.012 | -0.002 | -0.197 |
|---|
| IQGAP2 |   | 16 | 34 | 117 | 119 | 0.264 | -0.170 | -0.051 | -0.112 | 0.049 |
|---|
| LYAR |   | 1 | 652 | 1292 | 1295 | undef | 0.037 | -0.154 | undef | undef |
|---|
| APLP1 |   | 3 | 265 | 859 | 843 | -0.170 | 0.284 | 0.043 | -0.105 | -0.190 |
|---|
| RSL24D1 |   | 2 | 391 | 606 | 609 | 0.098 | 0.206 | -0.026 | 0.169 | -0.022 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| MYC |   | 24 | 21 | 148 | 137 | -0.056 | -0.101 | -0.169 | 0.188 | -0.134 |
|---|
| CTPS |   | 2 | 391 | 584 | 578 | -0.227 | 0.060 | -0.110 | 0.026 | -0.041 |
|---|
| BCL2 |   | 11 | 59 | 364 | 350 | 0.067 | -0.077 | -0.035 | 0.128 | 0.043 |
|---|
| S100A9 |   | 18 | 26 | 398 | 382 | 0.154 | 0.117 | 0.043 | -0.110 | -0.045 |
|---|
| RRAS2 |   | 6 | 120 | 563 | 549 | -0.089 | 0.155 | 0.076 | 0.289 | 0.031 |
|---|
| MPHOSPH6 |   | 8 | 84 | 756 | 726 | 0.217 | -0.084 | 0.175 | -0.269 | 0.146 |
|---|
| SP1 |   | 16 | 34 | 283 | 272 | -0.129 | 0.222 | -0.254 | 0.239 | 0.082 |
|---|
| CABYR |   | 1 | 652 | 1292 | 1295 | -0.221 | 0.015 | -0.003 | -0.046 | -0.088 |
|---|
| GSTM4 |   | 1 | 652 | 1292 | 1295 | 0.040 | -0.099 | -0.267 | 0.074 | -0.036 |
|---|
| ESRRG |   | 1 | 652 | 1292 | 1295 | 0.246 | -0.205 | 0.197 | -0.007 | -0.085 |
|---|
| PFKM |   | 4 | 198 | 492 | 499 | -0.193 | -0.030 | -0.079 | 0.293 | 0.118 |
|---|
| PDCD6 |   | 4 | 198 | 444 | 444 | undef | 0.091 | -0.060 | undef | undef |
|---|
| IKBKE |   | 8 | 84 | 756 | 726 | -0.033 | 0.227 | -0.041 | -0.095 | 0.079 |
|---|
| SYNCRIP |   | 1 | 652 | 1292 | 1295 | -0.216 | -0.087 | -0.018 | 0.037 | 0.119 |
|---|
| FMO2 |   | 1 | 652 | 1292 | 1295 | 0.118 | -0.041 | -0.094 | 0.174 | -0.046 |
|---|
| FRAT1 |   | 2 | 391 | 832 | 831 | -0.055 | -0.013 | -0.125 | 0.156 | 0.078 |
|---|
| TFDP1 |   | 4 | 198 | 339 | 341 | -0.194 | 0.123 | -0.178 | 0.129 | -0.065 |
|---|
| FHL1 |   | 3 | 265 | 1292 | 1278 | 0.136 | -0.041 | 0.104 | 0.301 | 0.266 |
|---|
| CDK2 |   | 16 | 34 | 129 | 124 | -0.104 | 0.196 | -0.096 | 0.109 | 0.362 |
|---|
| CBX5 |   | 2 | 391 | 1292 | 1279 | -0.061 | 0.155 | 0.016 | 0.148 | -0.009 |
|---|
| ESR1 |   | 4 | 198 | 1292 | 1277 | 0.046 | 0.026 | -0.002 | -0.053 | -0.062 |
|---|
| CLEC3B |   | 1 | 652 | 1292 | 1295 | 0.108 | 0.009 | -0.021 | 0.139 | 0.033 |
|---|
GO Enrichment output for subnetwork 1300 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of epidermis development | GO:0045684 |  | 1.474E-07 | 3.39E-04 |
|---|
| CTP metabolic process | GO:0046036 |  | 5.239E-07 | 6.025E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 7.043E-07 | 5.4E-04 |
|---|
| pyrimidine nucleoside triphosphate biosynthetic process | GO:0009148 |  | 8.677E-07 | 4.989E-04 |
|---|
| focal adhesion formation | GO:0048041 |  | 1.355E-06 | 6.232E-04 |
|---|
| pyrimidine nucleoside triphosphate metabolic process | GO:0009147 |  | 2.019E-06 | 7.741E-04 |
|---|
| regulation of epidermis development | GO:0045682 |  | 2.019E-06 | 6.635E-04 |
|---|
| pyrimidine ribonucleotide biosynthetic process | GO:0009220 |  | 4.032E-06 | 1.159E-03 |
|---|
| epidermis morphogenesis | GO:0048730 |  | 4.032E-06 | 1.03E-03 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 5.296E-06 | 1.218E-03 |
|---|
| pyrimidine ribonucleotide metabolic process | GO:0009218 |  | 5.463E-06 | 1.142E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of epidermis development | GO:0045684 |  | 8.136E-08 | 1.988E-04 |
|---|
| CTP metabolic process | GO:0046036 |  | 2.418E-07 | 2.954E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 2.661E-07 | 2.167E-04 |
|---|
| pyrimidine nucleoside triphosphate biosynthetic process | GO:0009148 |  | 3.783E-07 | 2.31E-04 |
|---|
| focal adhesion formation | GO:0048041 |  | 5.648E-07 | 2.759E-04 |
|---|
| pyrimidine nucleoside triphosphate metabolic process | GO:0009147 |  | 8.12E-07 | 3.306E-04 |
|---|
| regulation of epidermis development | GO:0045682 |  | 8.12E-07 | 2.834E-04 |
|---|
| pyrimidine ribonucleotide biosynthetic process | GO:0009220 |  | 1.536E-06 | 4.69E-04 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 2.015E-06 | 5.471E-04 |
|---|
| cellular glucose homeostasis | GO:0001678 |  | 2.015E-06 | 4.924E-04 |
|---|
| regulation of keratinocyte differentiation | GO:0045616 |  | 2.015E-06 | 4.476E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of epidermis development | GO:0045684 |  | 1.098E-07 | 2.643E-04 |
|---|
| CTP metabolic process | GO:0046036 |  | 1.967E-07 | 2.367E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 3.11E-07 | 2.494E-04 |
|---|
| pyrimidine nucleoside triphosphate biosynthetic process | GO:0009148 |  | 3.263E-07 | 1.962E-04 |
|---|
| focal adhesion formation | GO:0048041 |  | 7.614E-07 | 3.664E-04 |
|---|
| pyrimidine nucleoside triphosphate metabolic process | GO:0009147 |  | 7.614E-07 | 3.053E-04 |
|---|
| regulation of epidermis development | GO:0045682 |  | 1.094E-06 | 3.761E-04 |
|---|
| pyrimidine ribonucleotide biosynthetic process | GO:0009220 |  | 1.524E-06 | 4.585E-04 |
|---|
| pyrimidine ribonucleotide metabolic process | GO:0009218 |  | 2.068E-06 | 5.529E-04 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 2.527E-06 | 6.08E-04 |
|---|
| cellular glucose homeostasis | GO:0001678 |  | 2.527E-06 | 5.527E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of epidermis development | GO:0045684 |  | 1.474E-07 | 3.39E-04 |
|---|
| CTP metabolic process | GO:0046036 |  | 5.239E-07 | 6.025E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 7.043E-07 | 5.4E-04 |
|---|
| pyrimidine nucleoside triphosphate biosynthetic process | GO:0009148 |  | 8.677E-07 | 4.989E-04 |
|---|
| focal adhesion formation | GO:0048041 |  | 1.355E-06 | 6.232E-04 |
|---|
| pyrimidine nucleoside triphosphate metabolic process | GO:0009147 |  | 2.019E-06 | 7.741E-04 |
|---|
| regulation of epidermis development | GO:0045682 |  | 2.019E-06 | 6.635E-04 |
|---|
| pyrimidine ribonucleotide biosynthetic process | GO:0009220 |  | 4.032E-06 | 1.159E-03 |
|---|
| epidermis morphogenesis | GO:0048730 |  | 4.032E-06 | 1.03E-03 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 5.296E-06 | 1.218E-03 |
|---|
| pyrimidine ribonucleotide metabolic process | GO:0009218 |  | 5.463E-06 | 1.142E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of epidermis development | GO:0045684 |  | 1.474E-07 | 3.39E-04 |
|---|
| CTP metabolic process | GO:0046036 |  | 5.239E-07 | 6.025E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 7.043E-07 | 5.4E-04 |
|---|
| pyrimidine nucleoside triphosphate biosynthetic process | GO:0009148 |  | 8.677E-07 | 4.989E-04 |
|---|
| focal adhesion formation | GO:0048041 |  | 1.355E-06 | 6.232E-04 |
|---|
| pyrimidine nucleoside triphosphate metabolic process | GO:0009147 |  | 2.019E-06 | 7.741E-04 |
|---|
| regulation of epidermis development | GO:0045682 |  | 2.019E-06 | 6.635E-04 |
|---|
| pyrimidine ribonucleotide biosynthetic process | GO:0009220 |  | 4.032E-06 | 1.159E-03 |
|---|
| epidermis morphogenesis | GO:0048730 |  | 4.032E-06 | 1.03E-03 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 5.296E-06 | 1.218E-03 |
|---|
| pyrimidine ribonucleotide metabolic process | GO:0009218 |  | 5.463E-06 | 1.142E-03 |
|---|


