Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1289
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3380 | 1.220e-03 | 1.900e-05 | 1.481e-01 | 3.433e-09 |
|---|
| IPC | 0.3140 | 3.926e-01 | 2.186e-01 | 9.446e-01 | 8.106e-02 |
|---|
| Loi | 0.4901 | 2.900e-05 | 0.000e+00 | 4.680e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5943 | 1.000e-06 | 0.000e+00 | 1.249e-01 | 0.000e+00 |
|---|
| Wang | 0.2531 | 9.022e-03 | 7.354e-02 | 2.481e-01 | 1.646e-04 |
|---|
Expression data for subnetwork 1289 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
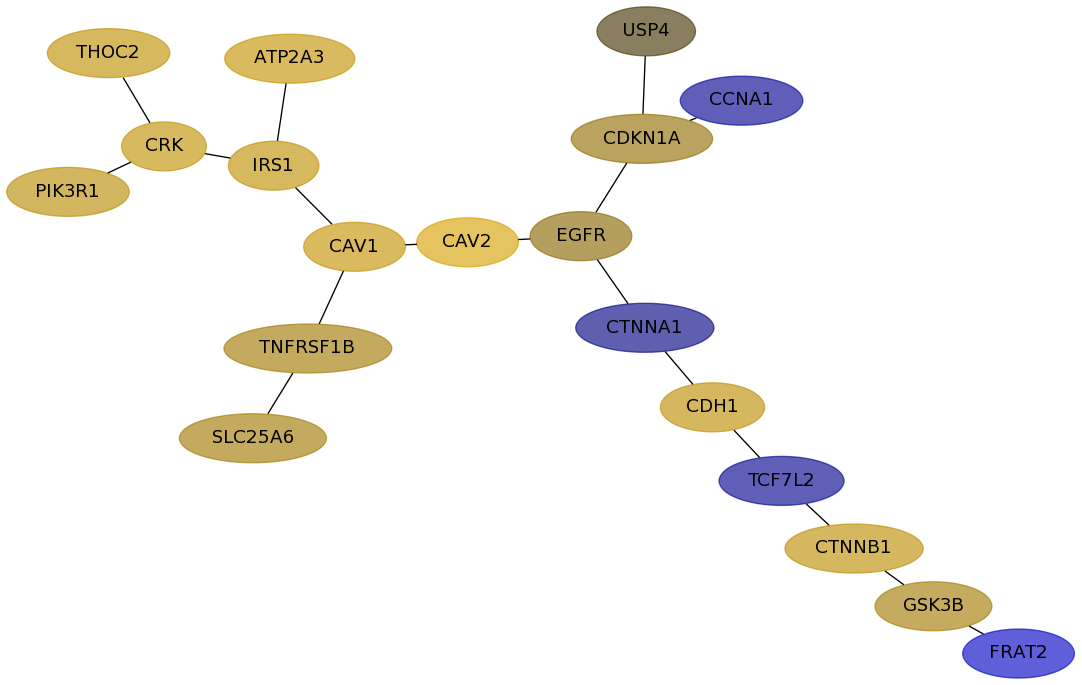
Score for each gene in subnetwork 1289 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| USP4 |   | 6 | 120 | 823 | 803 | 0.012 | -0.093 | 0.147 | 0.097 | 0.054 |
|---|
| CTNNA1 |   | 1 | 652 | 1218 | 1222 | -0.052 | -0.049 | 0.218 | 0.010 | -0.098 |
|---|
| ATP2A3 |   | 9 | 77 | 1 | 18 | 0.240 | -0.040 | 0.082 | -0.229 | -0.025 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| FRAT2 |   | 2 | 391 | 1218 | 1206 | -0.244 | 0.097 | -0.042 | 0.249 | 0.048 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| CAV1 |   | 10 | 67 | 439 | 428 | 0.251 | -0.092 | 0.124 | 0.079 | 0.119 |
|---|
| TCF7L2 |   | 17 | 30 | 271 | 264 | -0.071 | -0.021 | 0.143 | 0.280 | 0.101 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| TNFRSF1B |   | 12 | 55 | 563 | 544 | 0.109 | 0.024 | -0.187 | -0.096 | -0.106 |
|---|
| IRS1 |   | 14 | 42 | 1 | 14 | 0.217 | -0.219 | 0.065 | 0.165 | 0.064 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CRK |   | 27 | 17 | 206 | 193 | 0.232 | -0.116 | 0.106 | 0.090 | 0.017 |
|---|
| THOC2 |   | 11 | 59 | 419 | 406 | 0.217 | -0.094 | 0.119 | 0.068 | 0.072 |
|---|
| CCNA1 |   | 17 | 30 | 326 | 314 | -0.078 | 0.174 | 0.092 | 0.078 | -0.098 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| SLC25A6 |   | 3 | 265 | 778 | 770 | 0.111 | -0.178 | 0.115 | 0.040 | -0.080 |
|---|
GO Enrichment output for subnetwork 1289 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.181E-09 | 2.717E-06 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 8.028E-08 | 9.233E-05 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 8.702E-08 | 6.671E-05 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 8.702E-08 | 5.003E-05 |
|---|
| gas homeostasis | GO:0033483 |  | 1.738E-07 | 7.994E-05 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 1.738E-07 | 6.662E-05 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 1.738E-07 | 5.71E-05 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 3.037E-07 | 8.731E-05 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 4.852E-07 | 1.24E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 4.852E-07 | 1.116E-04 |
|---|
| mammary gland development | GO:0030879 |  | 5.128E-07 | 1.072E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 8.208E-15 | 2.005E-11 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 4.296E-08 | 5.248E-05 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 4.562E-08 | 3.715E-05 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 9.112E-08 | 5.565E-05 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 1.593E-07 | 7.782E-05 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 1.593E-07 | 6.485E-05 |
|---|
| positive regulation of insulin secretion | GO:0032024 |  | 2.545E-07 | 8.883E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.545E-07 | 7.773E-05 |
|---|
| gas homeostasis | GO:0033483 |  | 2.545E-07 | 6.909E-05 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 2.545E-07 | 6.218E-05 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 2.545E-07 | 5.653E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 6.545E-15 | 1.575E-11 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 5.079E-08 | 6.11E-05 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 6.073E-08 | 4.871E-05 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 1.213E-07 | 7.296E-05 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 1.213E-07 | 5.837E-05 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 2.12E-07 | 8.501E-05 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 2.12E-07 | 7.287E-05 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 2.12E-07 | 6.376E-05 |
|---|
| positive regulation of insulin secretion | GO:0032024 |  | 3.387E-07 | 9.056E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 3.387E-07 | 8.15E-05 |
|---|
| regulation of epithelial cell differentiation | GO:0030856 |  | 5.074E-07 | 1.11E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.181E-09 | 2.717E-06 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 8.028E-08 | 9.233E-05 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 8.702E-08 | 6.671E-05 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 8.702E-08 | 5.003E-05 |
|---|
| gas homeostasis | GO:0033483 |  | 1.738E-07 | 7.994E-05 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 1.738E-07 | 6.662E-05 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 1.738E-07 | 5.71E-05 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 3.037E-07 | 8.731E-05 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 4.852E-07 | 1.24E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 4.852E-07 | 1.116E-04 |
|---|
| mammary gland development | GO:0030879 |  | 5.128E-07 | 1.072E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.181E-09 | 2.717E-06 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 8.028E-08 | 9.233E-05 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 8.702E-08 | 6.671E-05 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 8.702E-08 | 5.003E-05 |
|---|
| gas homeostasis | GO:0033483 |  | 1.738E-07 | 7.994E-05 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 1.738E-07 | 6.662E-05 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 1.738E-07 | 5.71E-05 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 3.037E-07 | 8.731E-05 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 4.852E-07 | 1.24E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 4.852E-07 | 1.116E-04 |
|---|
| mammary gland development | GO:0030879 |  | 5.128E-07 | 1.072E-04 |
|---|


