Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1242
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3336 | 1.487e-03 | 2.600e-05 | 1.614e-01 | 6.239e-09 |
|---|
| IPC | 0.3311 | 3.647e-01 | 1.926e-01 | 9.389e-01 | 6.594e-02 |
|---|
| Loi | 0.4864 | 3.400e-05 | 0.000e+00 | 4.851e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5995 | 1.000e-06 | 0.000e+00 | 1.138e-01 | 0.000e+00 |
|---|
| Wang | 0.2559 | 8.073e-03 | 6.862e-02 | 2.358e-01 | 1.306e-04 |
|---|
Expression data for subnetwork 1242 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
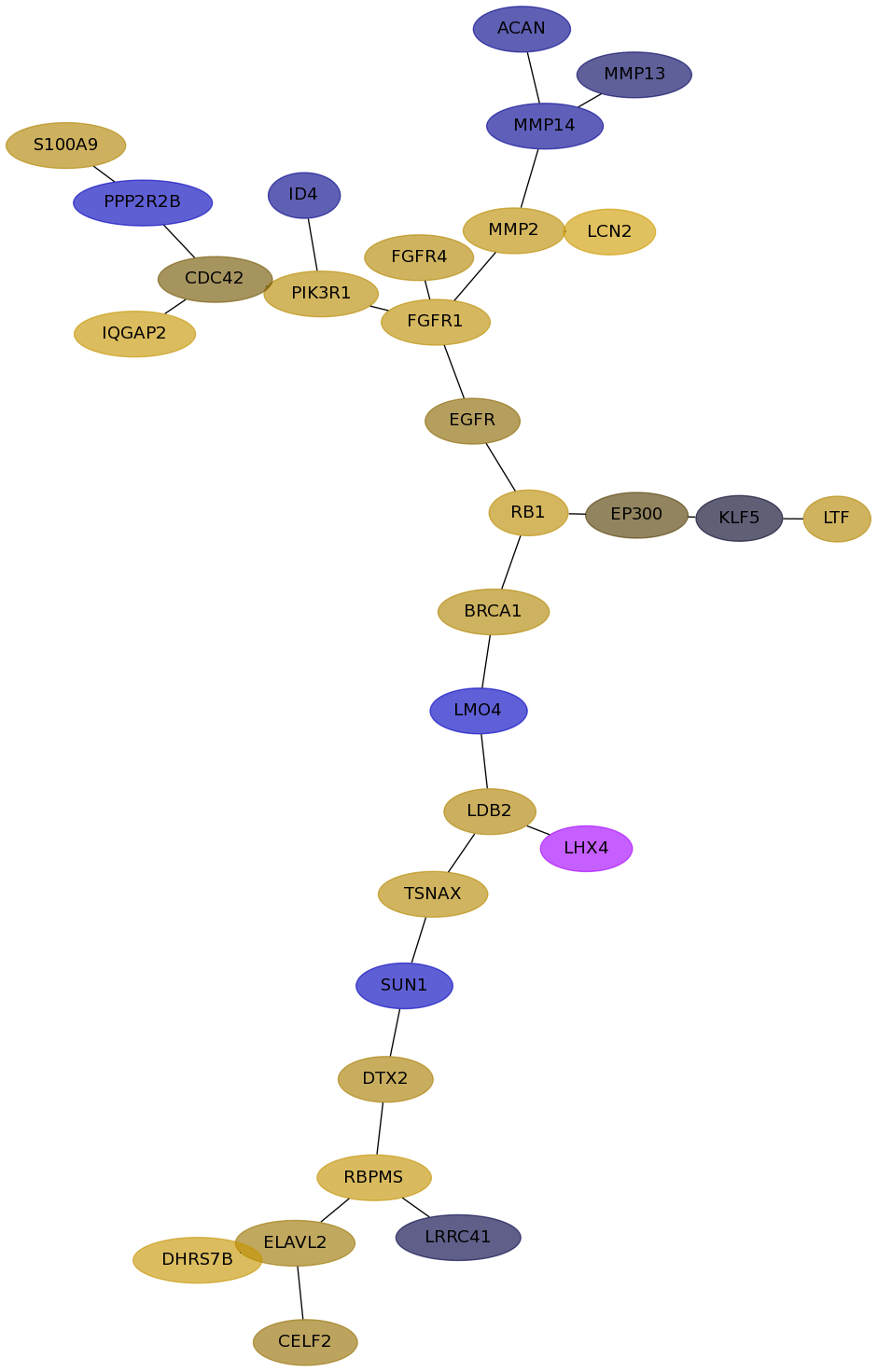
Score for each gene in subnetwork 1242 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| SUN1 |   | 1 | 652 | 797 | 810 | -0.211 | 0.079 | 0.153 | 0.054 | -0.069 |
|---|
| CELF2 |   | 3 | 265 | 634 | 620 | 0.083 | -0.124 | -0.086 | -0.056 | 0.002 |
|---|
| LCN2 |   | 6 | 120 | 797 | 779 | 0.316 | 0.113 | 0.145 | -0.098 | -0.284 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| ID4 |   | 13 | 47 | 444 | 430 | -0.060 | -0.179 | 0.137 | 0.241 | 0.036 |
|---|
| BRCA1 |   | 5 | 148 | 598 | 575 | 0.157 | -0.201 | -0.078 | -0.087 | 0.168 |
|---|
| LDB2 |   | 1 | 652 | 797 | 810 | 0.152 | -0.118 | 0.055 | 0.122 | 0.078 |
|---|
| LTF |   | 7 | 100 | 492 | 495 | 0.168 | -0.082 | 0.060 | -0.268 | -0.280 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| IQGAP2 |   | 16 | 34 | 117 | 119 | 0.264 | -0.170 | -0.051 | -0.112 | 0.049 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| ELAVL2 |   | 4 | 198 | 419 | 417 | 0.096 | 0.085 | 0.115 | 0.158 | -0.076 |
|---|
| TSNAX |   | 1 | 652 | 797 | 810 | 0.182 | 0.162 | 0.075 | 0.018 | 0.154 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| LHX4 |   | 1 | 652 | 797 | 810 | undef | -0.020 | 0.112 | undef | undef |
|---|
| LMO4 |   | 1 | 652 | 797 | 810 | -0.223 | -0.094 | 0.071 | 0.017 | -0.123 |
|---|
| MMP2 |   | 7 | 100 | 797 | 778 | 0.207 | -0.105 | 0.186 | -0.024 | 0.162 |
|---|
| ACAN |   | 4 | 198 | 797 | 792 | -0.060 | 0.156 | 0.132 | -0.024 | -0.083 |
|---|
| MMP14 |   | 2 | 391 | 57 | 80 | -0.077 | -0.114 | 0.056 | 0.011 | 0.148 |
|---|
| DTX2 |   | 2 | 391 | 419 | 433 | 0.130 | 0.005 | 0.174 | -0.122 | -0.200 |
|---|
| KLF5 |   | 25 | 19 | 57 | 50 | -0.005 | 0.077 | 0.117 | -0.007 | -0.006 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| RBPMS |   | 7 | 100 | 419 | 407 | 0.246 | 0.043 | 0.108 | 0.066 | -0.077 |
|---|
| MMP13 |   | 3 | 265 | 797 | 793 | -0.023 | 0.162 | 0.112 | -0.115 | 0.177 |
|---|
| S100A9 |   | 18 | 26 | 398 | 382 | 0.154 | 0.117 | 0.043 | -0.110 | -0.045 |
|---|
| LRRC41 |   | 3 | 265 | 419 | 419 | -0.012 | -0.136 | 0.210 | -0.080 | -0.125 |
|---|
| DHRS7B |   | 4 | 198 | 419 | 417 | 0.261 | 0.031 | -0.058 | -0.114 | 0.009 |
|---|
GO Enrichment output for subnetwork 1242 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.315E-09 | 1.223E-05 |
|---|
| nuclear migration | GO:0007097 |  | 4.437E-07 | 5.103E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 4.437E-07 | 3.402E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 8.852E-07 | 5.09E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.545E-06 | 7.107E-04 |
|---|
| regulation of lipid metabolic process | GO:0019216 |  | 1.714E-06 | 6.572E-04 |
|---|
| nucleus localization | GO:0051647 |  | 3.689E-06 | 1.212E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 5.257E-06 | 1.511E-03 |
|---|
| response to hypoxia | GO:0001666 |  | 6.008E-06 | 1.535E-03 |
|---|
| response to oxygen levels | GO:0070482 |  | 6.879E-06 | 1.582E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 7.209E-06 | 1.507E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 3.929E-09 | 9.599E-06 |
|---|
| nuclear migration | GO:0007097 |  | 4.166E-07 | 5.089E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 4.166E-07 | 3.393E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.162E-06 | 7.096E-04 |
|---|
| regulation of lipid metabolic process | GO:0019216 |  | 1.182E-06 | 5.777E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.739E-06 | 7.082E-04 |
|---|
| response to hypoxia | GO:0001666 |  | 3.279E-06 | 1.145E-03 |
|---|
| nucleus localization | GO:0051647 |  | 3.403E-06 | 1.039E-03 |
|---|
| response to oxygen levels | GO:0070482 |  | 3.7E-06 | 1.004E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 4.528E-06 | 1.106E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.445E-06 | 1.876E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.154E-09 | 5.184E-06 |
|---|
| nuclear migration | GO:0007097 |  | 4.468E-07 | 5.375E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 4.468E-07 | 3.584E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 7.803E-07 | 4.694E-04 |
|---|
| regulation of lipid metabolic process | GO:0019216 |  | 1.041E-06 | 5.011E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.246E-06 | 4.996E-04 |
|---|
| response to hypoxia | GO:0001666 |  | 3.646E-06 | 1.253E-03 |
|---|
| nucleus localization | GO:0051647 |  | 3.648E-06 | 1.097E-03 |
|---|
| response to oxygen levels | GO:0070482 |  | 4.113E-06 | 1.1E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 4.854E-06 | 1.168E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.227E-06 | 2.018E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.315E-09 | 1.223E-05 |
|---|
| nuclear migration | GO:0007097 |  | 4.437E-07 | 5.103E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 4.437E-07 | 3.402E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 8.852E-07 | 5.09E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.545E-06 | 7.107E-04 |
|---|
| regulation of lipid metabolic process | GO:0019216 |  | 1.714E-06 | 6.572E-04 |
|---|
| nucleus localization | GO:0051647 |  | 3.689E-06 | 1.212E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 5.257E-06 | 1.511E-03 |
|---|
| response to hypoxia | GO:0001666 |  | 6.008E-06 | 1.535E-03 |
|---|
| response to oxygen levels | GO:0070482 |  | 6.879E-06 | 1.582E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 7.209E-06 | 1.507E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.315E-09 | 1.223E-05 |
|---|
| nuclear migration | GO:0007097 |  | 4.437E-07 | 5.103E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 4.437E-07 | 3.402E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 8.852E-07 | 5.09E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.545E-06 | 7.107E-04 |
|---|
| regulation of lipid metabolic process | GO:0019216 |  | 1.714E-06 | 6.572E-04 |
|---|
| nucleus localization | GO:0051647 |  | 3.689E-06 | 1.212E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 5.257E-06 | 1.511E-03 |
|---|
| response to hypoxia | GO:0001666 |  | 6.008E-06 | 1.535E-03 |
|---|
| response to oxygen levels | GO:0070482 |  | 6.879E-06 | 1.582E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 7.209E-06 | 1.507E-03 |
|---|


