Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1236
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3334 | 1.501e-03 | 2.600e-05 | 1.620e-01 | 6.324e-09 |
|---|
| IPC | 0.3327 | 3.622e-01 | 1.903e-01 | 9.384e-01 | 6.468e-02 |
|---|
| Loi | 0.4858 | 3.500e-05 | 0.000e+00 | 4.879e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6001 | 1.000e-06 | 0.000e+00 | 1.126e-01 | 0.000e+00 |
|---|
| Wang | 0.2563 | 7.957e-03 | 6.800e-02 | 2.342e-01 | 1.267e-04 |
|---|
Expression data for subnetwork 1236 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
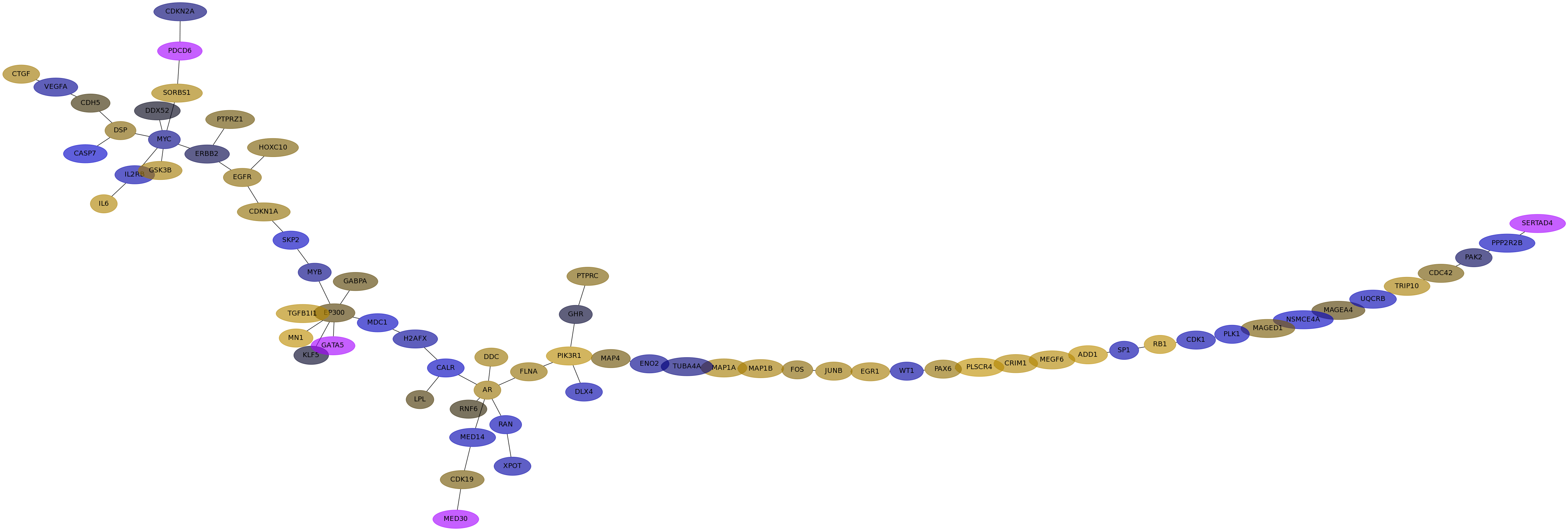
Score for each gene in subnetwork 1236 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| SORBS1 |   | 17 | 30 | 182 | 178 | 0.124 | -0.043 | 0.015 | 0.314 | 0.200 |
|---|
| PAX6 |   | 11 | 59 | 283 | 274 | 0.081 | 0.259 | -0.003 | 0.112 | 0.168 |
|---|
| ADD1 |   | 2 | 391 | 715 | 711 | 0.209 | -0.085 | 0.116 | 0.035 | -0.098 |
|---|
| UQCRB |   | 1 | 652 | 715 | 732 | -0.166 | 0.062 | -0.029 | 0.072 | 0.115 |
|---|
| TRIP10 |   | 3 | 265 | 715 | 705 | 0.127 | 0.012 | 0.154 | 0.049 | 0.144 |
|---|
| MAGED1 |   | 6 | 120 | 715 | 695 | 0.040 | -0.141 | 0.284 | -0.152 | 0.045 |
|---|
| ERBB2 |   | 2 | 391 | 715 | 711 | -0.014 | -0.103 | 0.092 | -0.090 | 0.046 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| MDC1 |   | 2 | 391 | 598 | 600 | -0.213 | -0.001 | 0.067 | 0.138 | 0.179 |
|---|
| CTGF |   | 10 | 67 | 57 | 52 | 0.106 | -0.169 | 0.181 | 0.126 | 0.209 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| RAN |   | 2 | 391 | 715 | 711 | -0.164 | 0.030 | -0.006 | 0.062 | undef |
|---|
| MED30 |   | 1 | 652 | 715 | 732 | undef | 0.185 | -0.225 | undef | undef |
|---|
| HOXC10 |   | 4 | 198 | 444 | 444 | 0.044 | -0.015 | 0.116 | -0.133 | -0.017 |
|---|
| TUBA4A |   | 3 | 265 | 715 | 705 | -0.041 | 0.157 | 0.136 | -0.010 | 0.156 |
|---|
| MEGF6 |   | 2 | 391 | 206 | 220 | 0.170 | 0.110 | 0.197 | 0.067 | 0.083 |
|---|
| PLK1 |   | 5 | 148 | 444 | 440 | -0.179 | 0.142 | -0.143 | 0.094 | -0.163 |
|---|
| GABPA |   | 6 | 120 | 206 | 200 | 0.019 | 0.033 | -0.183 | 0.066 | -0.161 |
|---|
| SERTAD4 |   | 7 | 100 | 1 | 20 | undef | 0.173 | 0.169 | undef | undef |
|---|
| FLNA |   | 7 | 100 | 206 | 198 | 0.077 | 0.081 | 0.107 | 0.148 | 0.126 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| DSP |   | 7 | 100 | 206 | 198 | 0.052 | 0.054 | 0.235 | -0.119 | 0.053 |
|---|
| CDK1 |   | 16 | 34 | 206 | 194 | -0.138 | 0.265 | -0.195 | 0.106 | 0.031 |
|---|
| XPOT |   | 1 | 652 | 715 | 732 | -0.123 | 0.126 | -0.014 | -0.044 | 0.134 |
|---|
| RNF6 |   | 1 | 652 | 715 | 732 | 0.006 | 0.109 | -0.128 | 0.002 | -0.018 |
|---|
| CDK19 |   | 1 | 652 | 715 | 732 | 0.036 | 0.171 | -0.043 | 0.090 | 0.054 |
|---|
| MAP4 |   | 1 | 652 | 715 | 732 | 0.029 | -0.115 | 0.217 | 0.221 | 0.268 |
|---|
| H2AFX |   | 4 | 198 | 117 | 125 | -0.086 | 0.111 | -0.072 | 0.121 | -0.010 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| LPL |   | 4 | 198 | 584 | 571 | 0.013 | -0.102 | -0.062 | 0.292 | -0.105 |
|---|
| KLF5 |   | 25 | 19 | 57 | 50 | -0.005 | 0.077 | 0.117 | -0.007 | -0.006 |
|---|
| MED14 |   | 1 | 652 | 715 | 732 | -0.157 | 0.091 | 0.000 | -0.014 | 0.105 |
|---|
| GATA5 |   | 1 | 652 | 715 | 732 | undef | 0.161 | -0.031 | undef | undef |
|---|
| TGFB1I1 |   | 16 | 34 | 326 | 315 | 0.176 | -0.105 | 0.229 | 0.143 | 0.298 |
|---|
| NSMCE4A |   | 3 | 265 | 715 | 705 | -0.228 | 0.177 | -0.116 | 0.290 | -0.091 |
|---|
| PLSCR4 |   | 2 | 391 | 444 | 458 | 0.223 | -0.069 | 0.140 | 0.231 | 0.212 |
|---|
| MAGEA4 |   | 1 | 652 | 715 | 732 | 0.017 | undef | -0.083 | 0.236 | -0.005 |
|---|
| EGR1 |   | 11 | 59 | 283 | 274 | 0.118 | -0.184 | 0.259 | 0.130 | 0.122 |
|---|
| ENO2 |   | 2 | 391 | 206 | 220 | -0.066 | 0.085 | 0.114 | 0.148 | 0.208 |
|---|
| CALR |   | 10 | 67 | 117 | 121 | -0.216 | -0.105 | 0.002 | -0.061 | -0.075 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| MAP1A |   | 1 | 652 | 715 | 732 | 0.121 | -0.045 | 0.143 | -0.054 | 0.147 |
|---|
| MYC |   | 24 | 21 | 148 | 137 | -0.056 | -0.101 | -0.169 | 0.188 | -0.134 |
|---|
| GHR |   | 14 | 42 | 148 | 146 | -0.006 | -0.112 | 0.174 | 0.090 | 0.201 |
|---|
| JUNB |   | 5 | 148 | 696 | 672 | 0.101 | 0.057 | 0.086 | 0.065 | -0.015 |
|---|
| PAK2 |   | 20 | 24 | 1 | 11 | -0.019 | 0.014 | 0.161 | 0.081 | 0.062 |
|---|
| PTPRZ1 |   | 1 | 652 | 715 | 732 | 0.029 | -0.114 | 0.183 | 0.233 | -0.095 |
|---|
| WT1 |   | 8 | 84 | 283 | 278 | -0.106 | 0.254 | -0.039 | 0.036 | 0.172 |
|---|
| DLX4 |   | 2 | 391 | 715 | 711 | -0.133 | 0.291 | 0.057 | -0.004 | -0.059 |
|---|
| DDC |   | 4 | 198 | 715 | 700 | 0.093 | -0.121 | 0.157 | -0.078 | 0.114 |
|---|
| SP1 |   | 16 | 34 | 283 | 272 | -0.129 | 0.222 | -0.254 | 0.239 | 0.082 |
|---|
| MYB |   | 3 | 265 | 148 | 166 | -0.054 | -0.028 | 0.075 | 0.002 | -0.012 |
|---|
| MN1 |   | 6 | 120 | 148 | 156 | 0.213 | -0.166 | -0.047 | 0.150 | 0.011 |
|---|
| CRIM1 |   | 1 | 652 | 715 | 732 | 0.162 | -0.134 | 0.162 | 0.122 | 0.180 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| SKP2 |   | 14 | 42 | 148 | 146 | -0.228 | 0.092 | -0.043 | 0.237 | -0.009 |
|---|
| IL2RB |   | 2 | 391 | 715 | 711 | -0.133 | 0.079 | -0.240 | -0.175 | -0.068 |
|---|
| FOS |   | 20 | 24 | 148 | 138 | 0.060 | -0.137 | 0.129 | 0.185 | 0.234 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| MAP1B |   | 13 | 47 | 148 | 148 | 0.118 | -0.123 | 0.263 | -0.107 | 0.100 |
|---|
| PTPRC |   | 5 | 148 | 715 | 697 | 0.043 | -0.031 | -0.204 | -0.050 | -0.068 |
|---|
| CASP7 |   | 8 | 84 | 283 | 278 | -0.270 | 0.216 | 0.095 | 0.255 | 0.035 |
|---|
| PDCD6 |   | 4 | 198 | 444 | 444 | undef | 0.091 | -0.060 | undef | undef |
|---|
| CDH5 |   | 2 | 391 | 364 | 375 | 0.009 | -0.159 | 0.023 | 0.180 | 0.033 |
|---|
| IL6 |   | 3 | 265 | 715 | 705 | 0.155 | 0.257 | -0.106 | 0.226 | 0.035 |
|---|
| AR |   | 9 | 77 | 283 | 277 | 0.089 | -0.163 | 0.081 | -0.035 | -0.030 |
|---|
| CDKN2A |   | 10 | 67 | 364 | 351 | -0.037 | 0.263 | -0.113 | 0.111 | -0.105 |
|---|
| DDX52 |   | 1 | 652 | 715 | 732 | -0.003 | 0.070 | -0.095 | 0.053 | 0.118 |
|---|
GO Enrichment output for subnetwork 1236 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mitochondrial electron transport. ubiquinol to cytochrome c | GO:0006122 |  | 3.816E-08 | 8.777E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 8.855E-08 | 1.018E-04 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 5.224E-07 | 4.005E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.094E-07 | 3.504E-04 |
|---|
| regulation of mast cell proliferation | GO:0070666 |  | 7.015E-07 | 3.227E-04 |
|---|
| regulation of leukocyte proliferation | GO:0070663 |  | 1.048E-06 | 4.019E-04 |
|---|
| regulation of B cell activation | GO:0050864 |  | 1.545E-06 | 5.077E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.341E-06 | 9.605E-04 |
|---|
| nuclear migration | GO:0007097 |  | 3.617E-06 | 9.245E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.617E-06 | 8.32E-04 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 3.626E-06 | 7.581E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mitochondrial electron transport. ubiquinol to cytochrome c | GO:0006122 |  | 6.999E-09 | 1.71E-05 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 2.025E-08 | 2.474E-05 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 2.748E-08 | 2.238E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 3.242E-08 | 1.98E-05 |
|---|
| intracellular receptor-mediated signaling pathway | GO:0030522 |  | 4.601E-08 | 2.248E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.88E-08 | 3.208E-05 |
|---|
| regulation of mast cell proliferation | GO:0070666 |  | 1.163E-07 | 4.058E-05 |
|---|
| regulation of leukocyte proliferation | GO:0070663 |  | 1.88E-07 | 5.74E-05 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 2.259E-07 | 6.133E-05 |
|---|
| regulation of B cell activation | GO:0050864 |  | 2.794E-07 | 6.825E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 9.224E-07 | 2.049E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mitochondrial electron transport. ubiquinol to cytochrome c | GO:0006122 |  | 1.004E-08 | 2.416E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.334E-08 | 2.808E-05 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 2.891E-08 | 2.318E-05 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 3.627E-08 | 2.181E-05 |
|---|
| intracellular receptor-mediated signaling pathway | GO:0030522 |  | 6.219E-08 | 2.993E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.332E-07 | 5.343E-05 |
|---|
| regulation of mast cell proliferation | GO:0070666 |  | 1.731E-07 | 5.949E-05 |
|---|
| regulation of leukocyte proliferation | GO:0070663 |  | 2.508E-07 | 7.543E-05 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 3.235E-07 | 8.648E-05 |
|---|
| regulation of B cell activation | GO:0050864 |  | 4.347E-07 | 1.046E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.299E-06 | 2.841E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mitochondrial electron transport. ubiquinol to cytochrome c | GO:0006122 |  | 3.816E-08 | 8.777E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 8.855E-08 | 1.018E-04 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 5.224E-07 | 4.005E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.094E-07 | 3.504E-04 |
|---|
| regulation of mast cell proliferation | GO:0070666 |  | 7.015E-07 | 3.227E-04 |
|---|
| regulation of leukocyte proliferation | GO:0070663 |  | 1.048E-06 | 4.019E-04 |
|---|
| regulation of B cell activation | GO:0050864 |  | 1.545E-06 | 5.077E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.341E-06 | 9.605E-04 |
|---|
| nuclear migration | GO:0007097 |  | 3.617E-06 | 9.245E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.617E-06 | 8.32E-04 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 3.626E-06 | 7.581E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mitochondrial electron transport. ubiquinol to cytochrome c | GO:0006122 |  | 3.816E-08 | 8.777E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 8.855E-08 | 1.018E-04 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 5.224E-07 | 4.005E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.094E-07 | 3.504E-04 |
|---|
| regulation of mast cell proliferation | GO:0070666 |  | 7.015E-07 | 3.227E-04 |
|---|
| regulation of leukocyte proliferation | GO:0070663 |  | 1.048E-06 | 4.019E-04 |
|---|
| regulation of B cell activation | GO:0050864 |  | 1.545E-06 | 5.077E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.341E-06 | 9.605E-04 |
|---|
| nuclear migration | GO:0007097 |  | 3.617E-06 | 9.245E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.617E-06 | 8.32E-04 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 3.626E-06 | 7.581E-04 |
|---|


